| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 10605792 | Carbohydrate Research | 2010 | 4 Pages |
Abstract
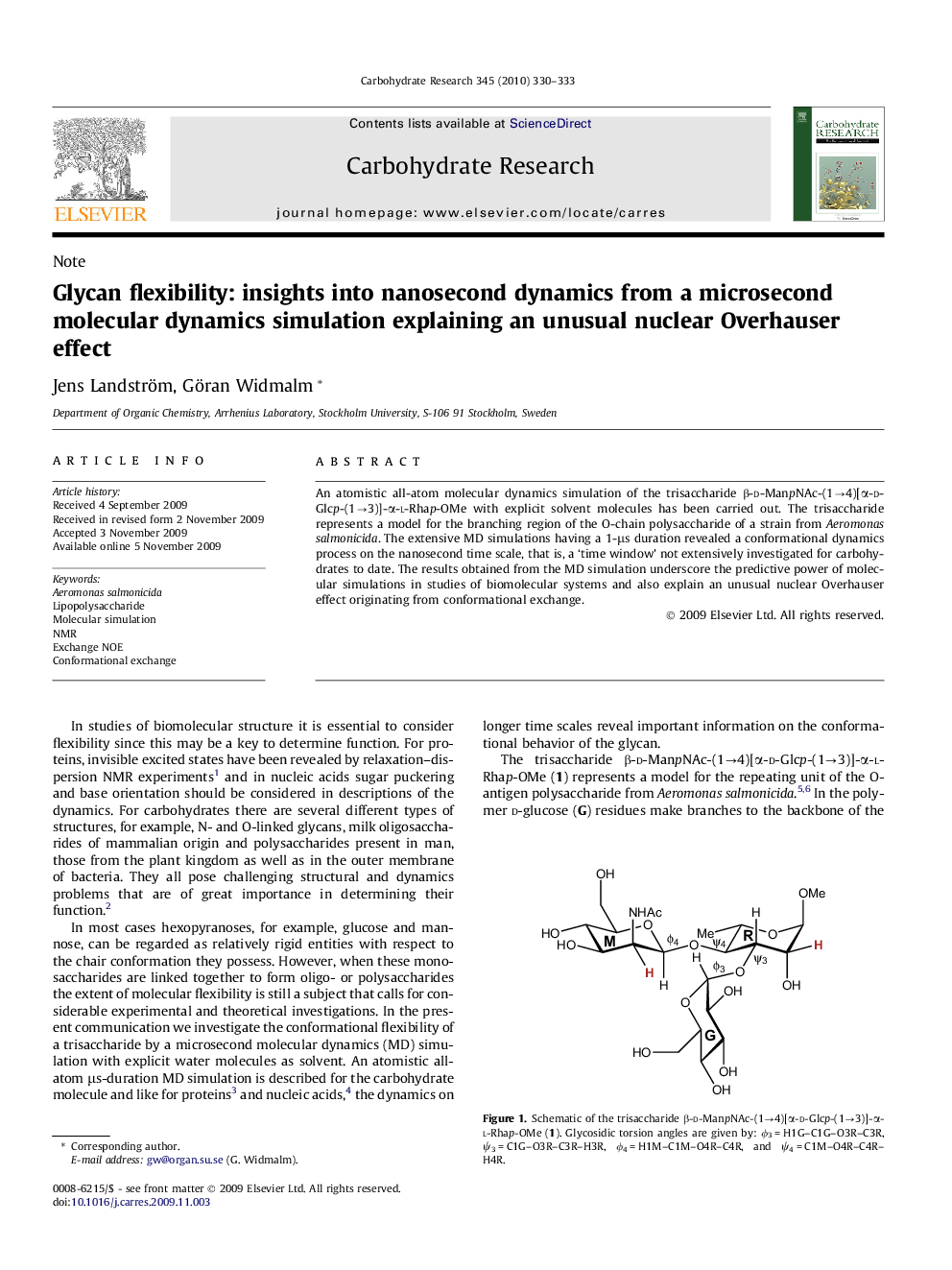
An atomistic all-atom molecular dynamics simulation of the trisaccharide β-d-ManpNAc-(1â4)[α-d-Glcp-(1â3)]-α-l-Rhap-OMe with explicit solvent molecules has been carried out. The trisaccharide represents a model for the branching region of the O-chain polysaccharide of a strain from Aeromonas salmonicida. The extensive MD simulations having a 1-μs duration revealed a conformational dynamics process on the nanosecond time scale, that is, a 'time window' not extensively investigated for carbohydrates to date. The results obtained from the MD simulation underscore the predictive power of molecular simulations in studies of biomolecular systems and also explain an unusual nuclear Overhauser effect originating from conformational exchange.
Related Topics
Physical Sciences and Engineering
Chemistry
Organic Chemistry
Authors
Jens Landström, Göran Widmalm,