| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 2035062 | Cell | 2016 | 13 Pages |
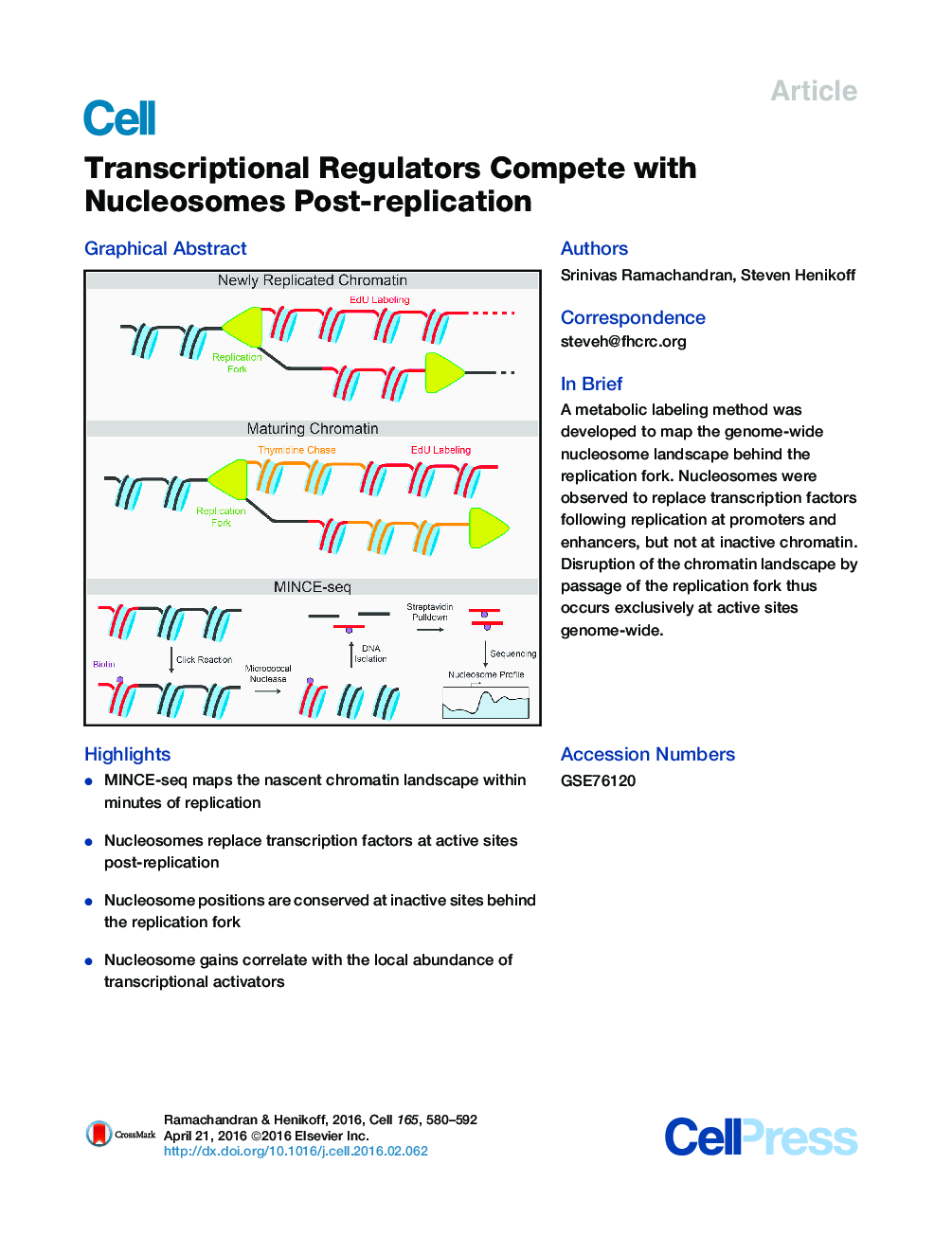
•MINCE-seq maps the nascent chromatin landscape within minutes of replication•Nucleosomes replace transcription factors at active sites post-replication•Nucleosome positions are conserved at inactive sites behind the replication fork•Nucleosome gains correlate with the local abundance of transcriptional activators
SummaryEvery nucleosome across the genome must be disrupted and reformed when the replication fork passes, but how chromatin organization is re-established following replication is unknown. To address this problem, we have developed Mapping In vivo Nascent Chromatin with EdU and sequencing (MINCE-seq) to characterize the genome-wide location of nucleosomes and other chromatin proteins behind replication forks at high temporal and spatial resolution. We find that the characteristic chromatin landscape at Drosophila promoters and enhancers is lost upon replication. The most conspicuous changes are at promoters that have high levels of RNA polymerase II (RNAPII) stalling and DNA accessibility and show specific enrichment for the BRM remodeler. Enhancer chromatin is also disrupted during replication, suggesting a role for transcription factor (TF) competition in nucleosome re-establishment. Thus, the characteristic nucleosome landscape emerges from a uniformly packaged genome by the action of TFs, RNAPII, and remodelers minutes after replication fork passage.
Graphical AbstractFigure optionsDownload full-size imageDownload high-quality image (238 K)Download as PowerPoint slide