| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 2077305 | Cell Stem Cell | 2015 | 14 Pages |
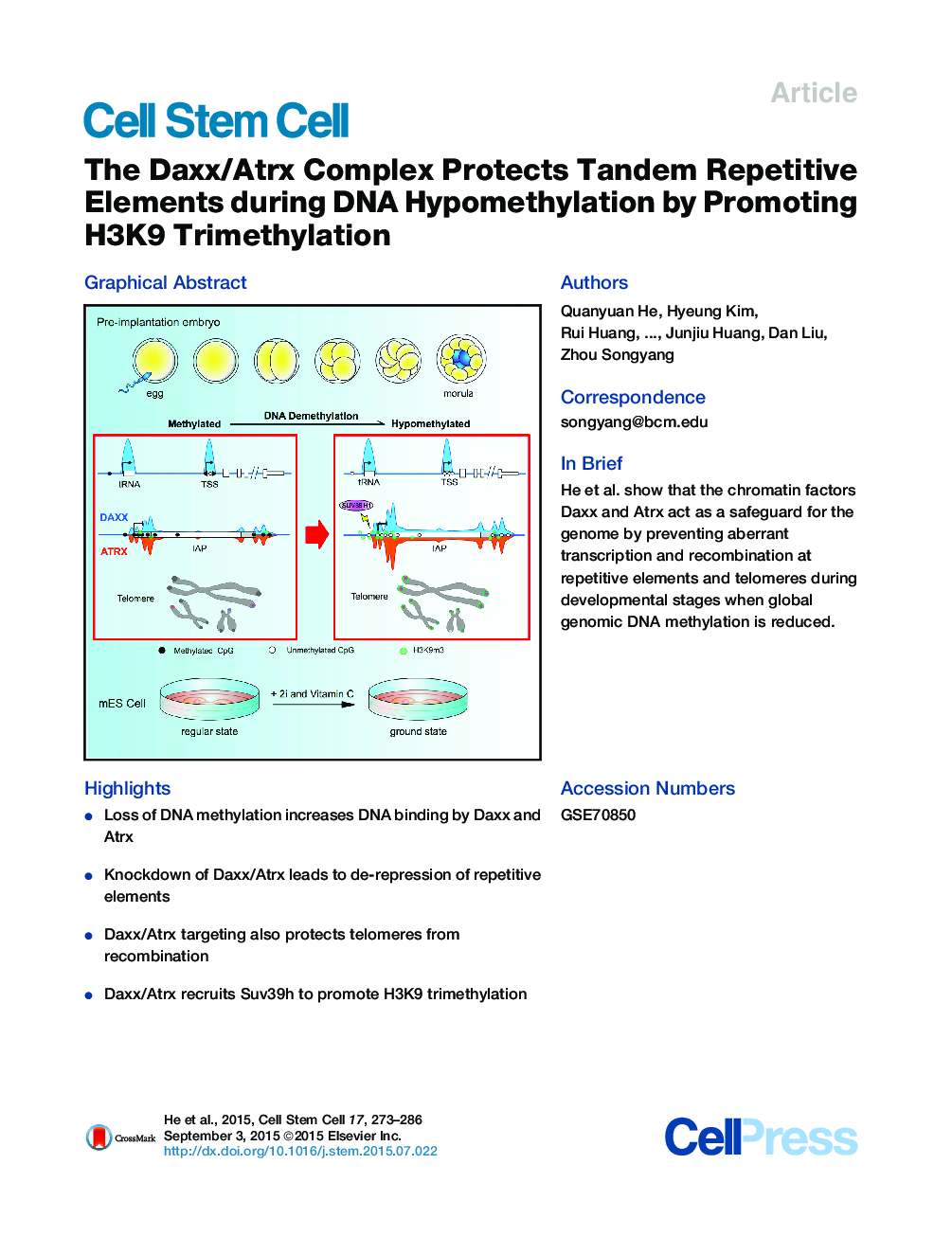
•Loss of DNA methylation increases DNA binding by Daxx and Atrx•Knockdown of Daxx/Atrx leads to de-repression of repetitive elements•Daxx/Atrx targeting also protects telomeres from recombination•Daxx/Atrx recruits Suv39h to promote H3K9 trimethylation
SummaryIn mammals, DNA methylation is essential for protecting repetitive sequences from aberrant transcription and recombination. In some developmental contexts (e.g., preimplantation embryos) DNA is hypomethylated but repetitive elements are not dysregulated, suggesting that alternative protection mechanisms exist. Here we explore the processes involved by investigating the role of the chromatin factors Daxx and Atrx. Using genome-wide binding and transcriptome analysis, we found that Daxx and Atrx have distinct chromatin-binding profiles and are co-enriched at tandem repetitive elements in wild-type mouse ESCs. Global DNA hypomethylation further promoted recruitment of the Daxx/Atrx complex to tandem repeat sequences, including retrotransposons and telomeres. Knockdown of Daxx/Atrx in cells with hypomethylated genomes exacerbated aberrant transcriptional de-repression of repeat elements and telomere dysfunction. Mechanistically, Daxx/Atrx-mediated repression seems to involve Suv39h recruitment and H3K9 trimethylation. Our data therefore suggest that Daxx and Atrx safeguard the genome by silencing repetitive elements when DNA methylation levels are low.
Graphical AbstractFigure optionsDownload full-size imageDownload high-quality image (242 K)Download as PowerPoint slide