| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 2792561 | Cell Metabolism | 2015 | 12 Pages |
•4,698 deletions tested yields the most comprehensive yeast data set on aging•Longevity clusters center on known, conserved biological processes•Enrichment of lifespan-extending C. elegans orthologs suggests conservation•Genome-wide information uncovered aging pathways such as tRNA transport
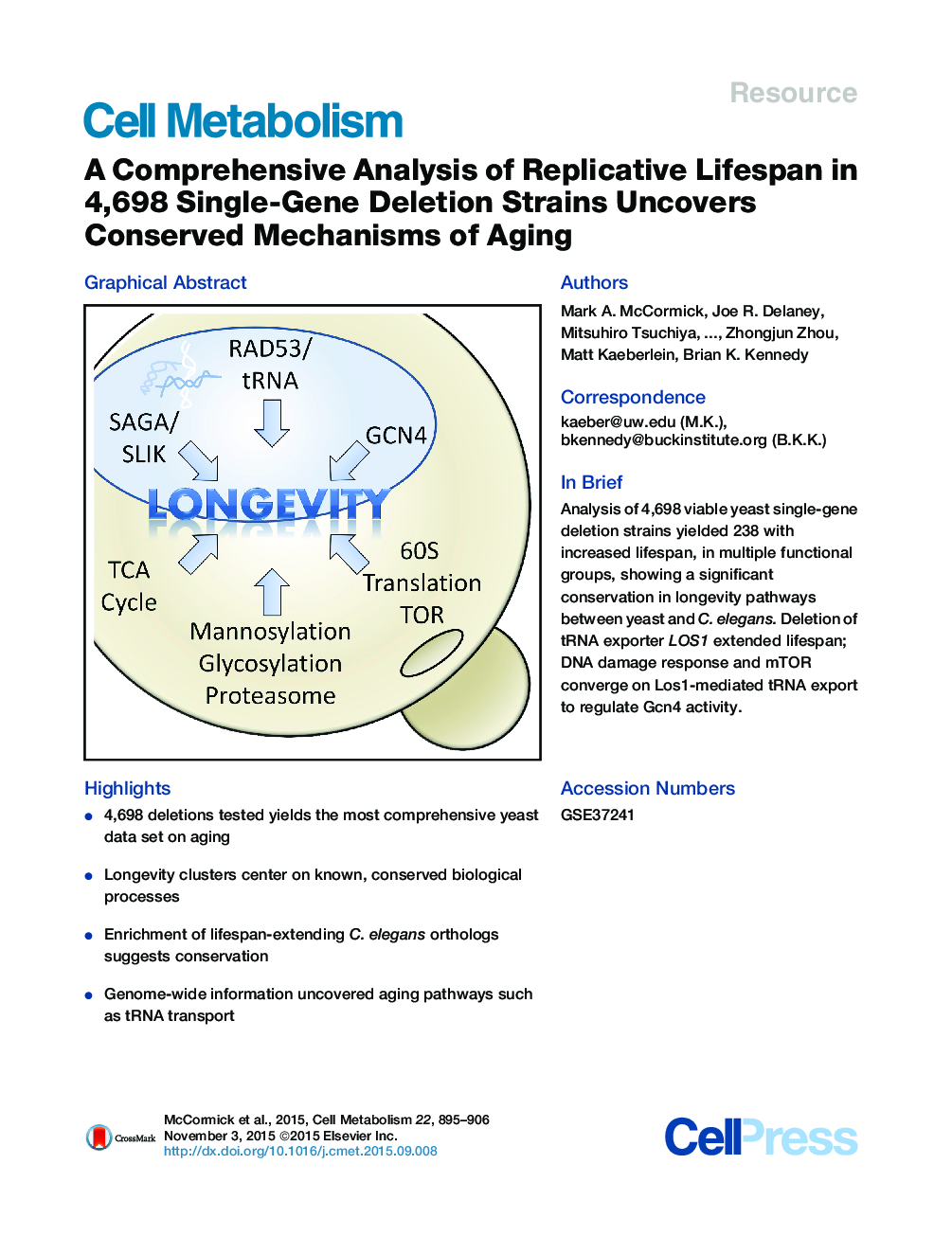
SummaryMany genes that affect replicative lifespan (RLS) in the budding yeast Saccharomyces cerevisiae also affect aging in other organisms such as C. elegans and M. musculus. We performed a systematic analysis of yeast RLS in a set of 4,698 viable single-gene deletion strains. Multiple functional gene clusters were identified, and full genome-to-genome comparison demonstrated a significant conservation in longevity pathways between yeast and C. elegans. Among the mechanisms of aging identified, deletion of tRNA exporter LOS1 robustly extended lifespan. Dietary restriction (DR) and inhibition of mechanistic Target of Rapamycin (mTOR) exclude Los1 from the nucleus in a Rad53-dependent manner. Moreover, lifespan extension from deletion of LOS1 is nonadditive with DR or mTOR inhibition, and results in Gcn4 transcription factor activation. Thus, the DNA damage response and mTOR converge on Los1-mediated nuclear tRNA export to regulate Gcn4 activity and aging.
Graphical AbstractFigure optionsDownload full-size imageDownload high-quality image (214 K)Download as PowerPoint slide