| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 4997607 | Bioresource Technology | 2017 | 9 Pages |
â¢First study on identification of autotrophic denitrifying communities by DNA-SIP.â¢High efficient NO3â-N, TN removals from organic-free influent based on CEAD process.â¢Strongly labeled nirS gene indicating H13CO3â assimilated by autotrophic denitrifiers.â¢Thiobacillus was the most dominant autotrophic denitrifiers though sulfur was absent.
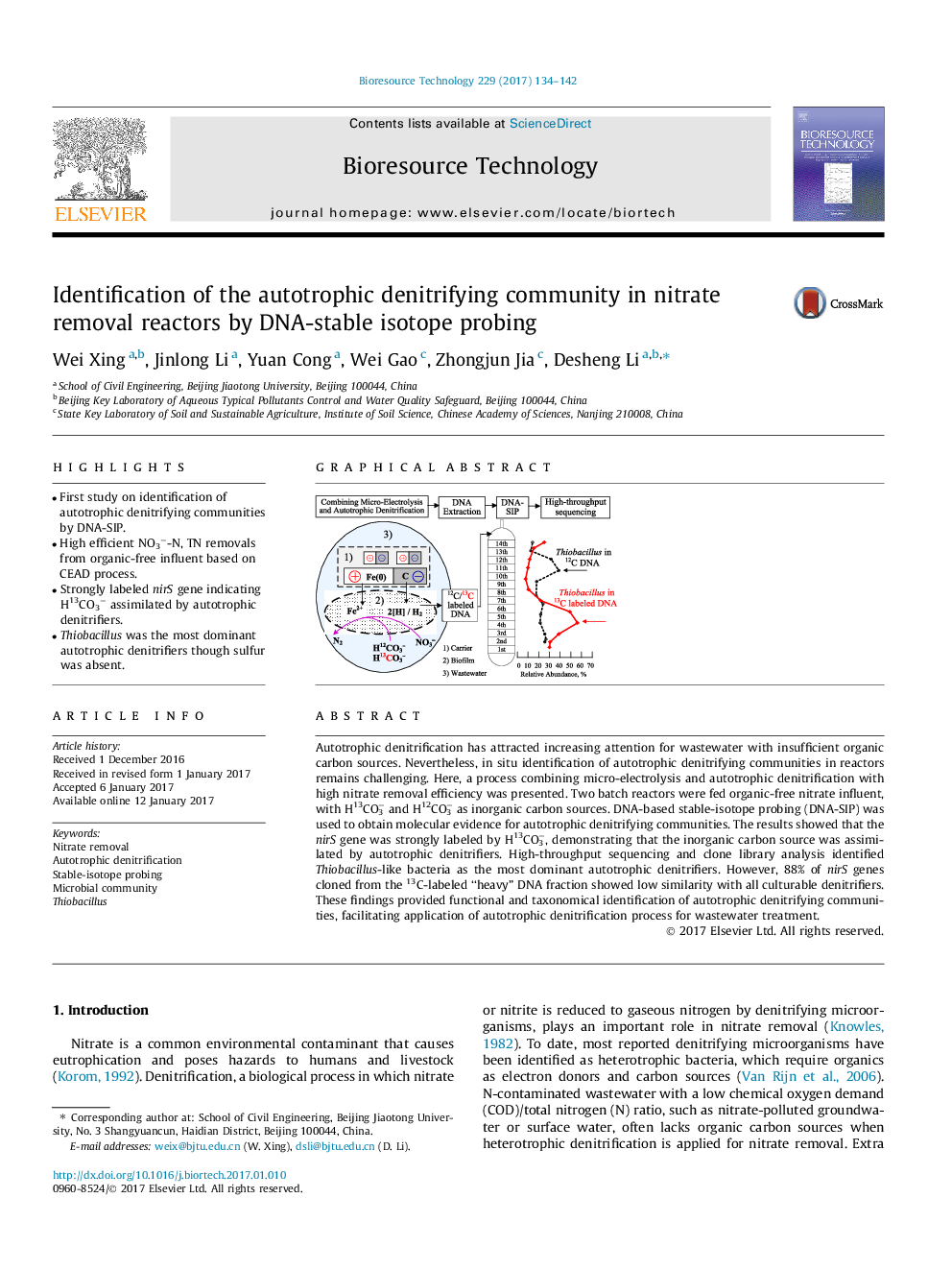
Autotrophic denitrification has attracted increasing attention for wastewater with insufficient organic carbon sources. Nevertheless, in situ identification of autotrophic denitrifying communities in reactors remains challenging. Here, a process combining micro-electrolysis and autotrophic denitrification with high nitrate removal efficiency was presented. Two batch reactors were fed organic-free nitrate influent, with H13CO3â and H12CO3â as inorganic carbon sources. DNA-based stable-isotope probing (DNA-SIP) was used to obtain molecular evidence for autotrophic denitrifying communities. The results showed that the nirS gene was strongly labeled by H13CO3â, demonstrating that the inorganic carbon source was assimilated by autotrophic denitrifiers. High-throughput sequencing and clone library analysis identified Thiobacillus-like bacteria as the most dominant autotrophic denitrifiers. However, 88% of nirS genes cloned from the 13C-labeled “heavy” DNA fraction showed low similarity with all culturable denitrifiers. These findings provided functional and taxonomical identification of autotrophic denitrifying communities, facilitating application of autotrophic denitrification process for wastewater treatment.
Graphical abstractDownload high-res image (228KB)Download full-size image