| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 5371161 | Biophysical Chemistry | 2013 | 10 Pages |
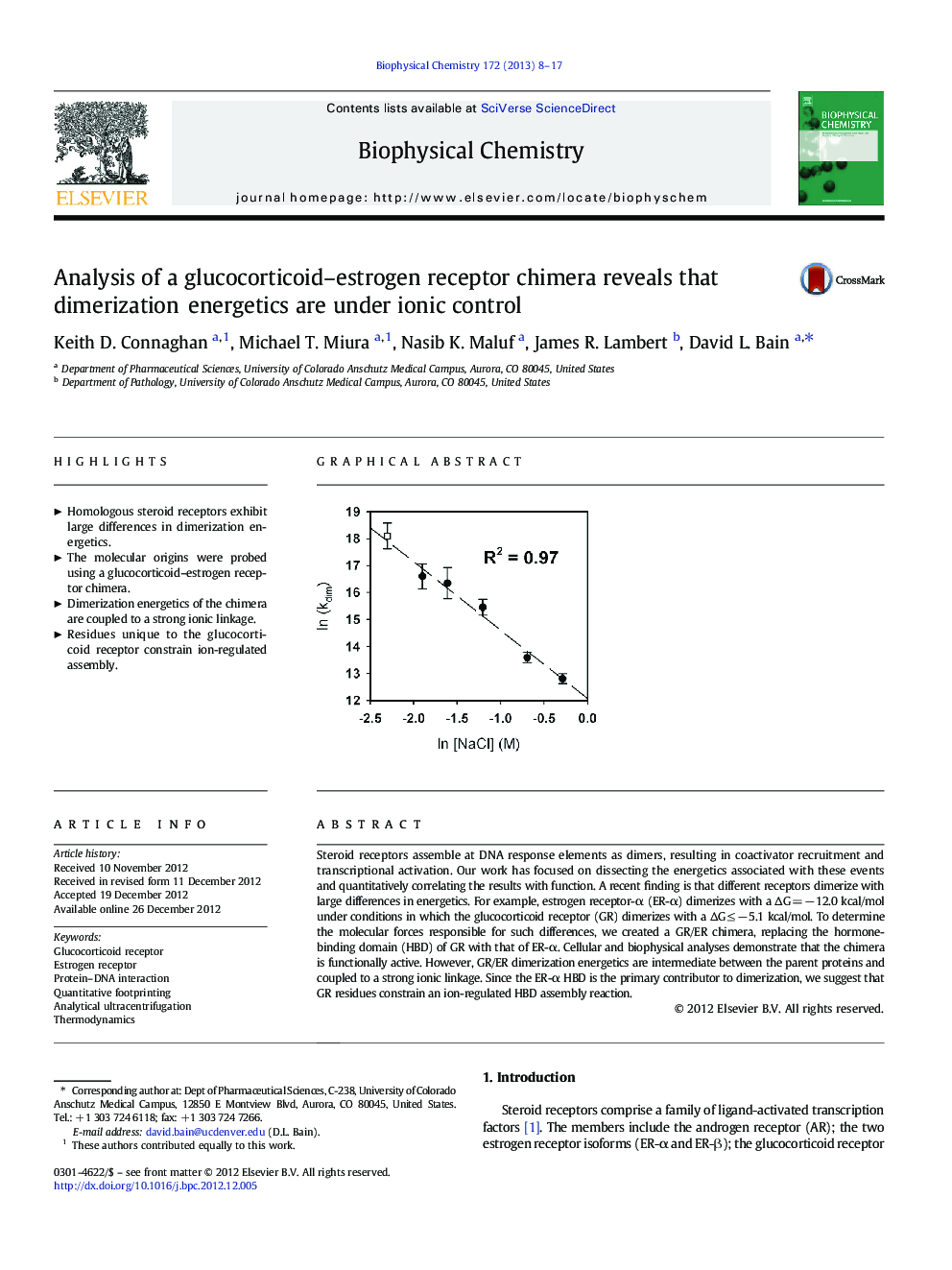
Steroid receptors assemble at DNA response elements as dimers, resulting in coactivator recruitment and transcriptional activation. Our work has focused on dissecting the energetics associated with these events and quantitatively correlating the results with function. A recent finding is that different receptors dimerize with large differences in energetics. For example, estrogen receptor-α (ER-α) dimerizes with a ÎG = â 12.0 kcal/mol under conditions in which the glucocorticoid receptor (GR) dimerizes with a ÎG â¤Â â 5.1 kcal/mol. To determine the molecular forces responsible for such differences, we created a GR/ER chimera, replacing the hormone-binding domain (HBD) of GR with that of ER-α. Cellular and biophysical analyses demonstrate that the chimera is functionally active. However, GR/ER dimerization energetics are intermediate between the parent proteins and coupled to a strong ionic linkage. Since the ER-α HBD is the primary contributor to dimerization, we suggest that GR residues constrain an ion-regulated HBD assembly reaction.
Graphical abstractDownload full-size imageHighlights⺠Homologous steroid receptors exhibit large differences in dimerization energetics. ⺠The molecular origins were probed using a glucocorticoid-estrogen receptor chimera. ⺠Dimerization energetics of the chimera are coupled to a strong ionic linkage. ⺠Residues unique to the glucocorticoid receptor constrain ion-regulated assembly.