| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 1166100 | Analytica Chimica Acta | 2012 | 5 Pages |
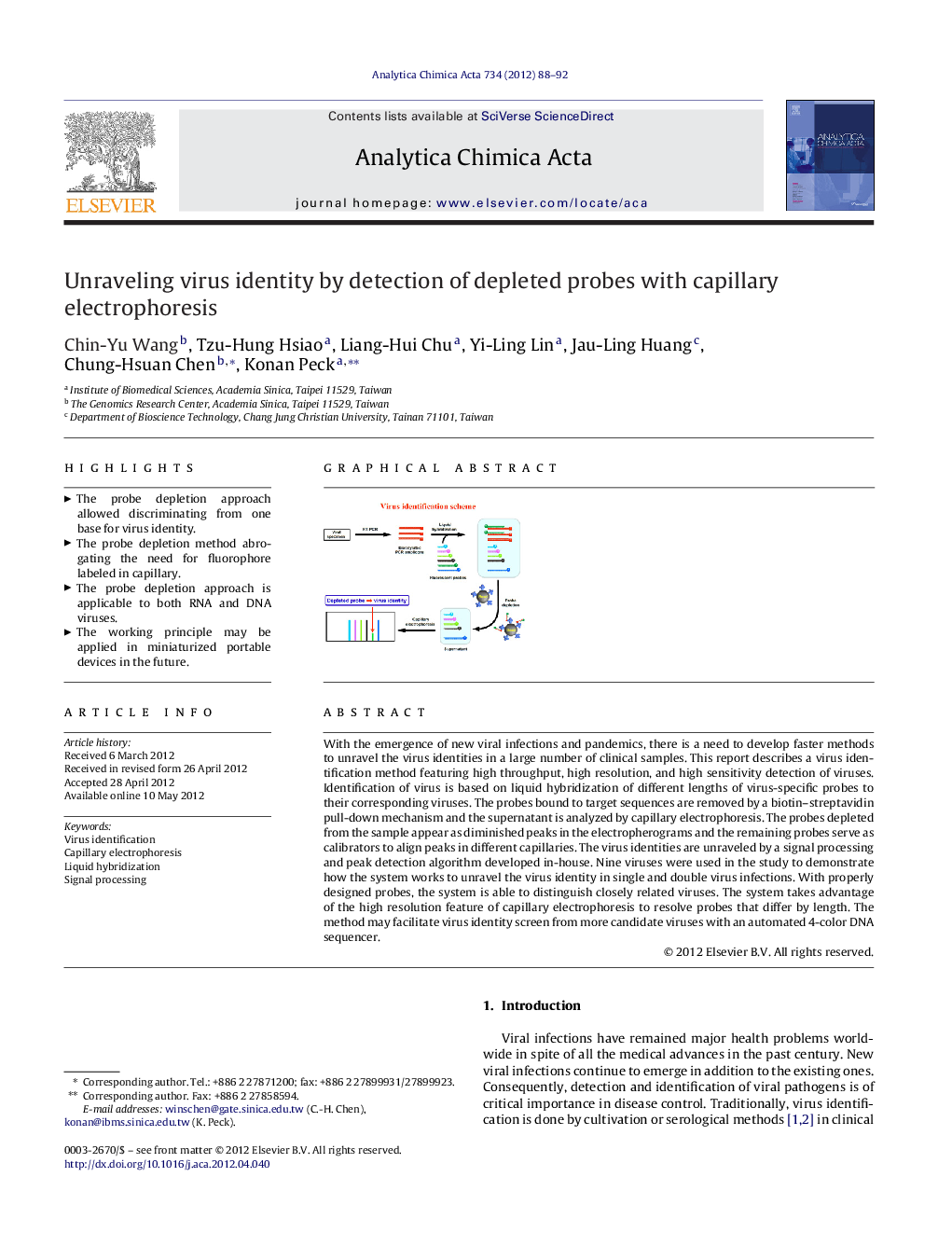
With the emergence of new viral infections and pandemics, there is a need to develop faster methods to unravel the virus identities in a large number of clinical samples. This report describes a virus identification method featuring high throughput, high resolution, and high sensitivity detection of viruses. Identification of virus is based on liquid hybridization of different lengths of virus-specific probes to their corresponding viruses. The probes bound to target sequences are removed by a biotin–streptavidin pull-down mechanism and the supernatant is analyzed by capillary electrophoresis. The probes depleted from the sample appear as diminished peaks in the electropherograms and the remaining probes serve as calibrators to align peaks in different capillaries. The virus identities are unraveled by a signal processing and peak detection algorithm developed in-house. Nine viruses were used in the study to demonstrate how the system works to unravel the virus identity in single and double virus infections. With properly designed probes, the system is able to distinguish closely related viruses. The system takes advantage of the high resolution feature of capillary electrophoresis to resolve probes that differ by length. The method may facilitate virus identity screen from more candidate viruses with an automated 4-color DNA sequencer.
Graphical abstractFigure optionsDownload full-size imageDownload as PowerPoint slideHighlights► The probe depletion approach allowed discriminating from one base for virus identity. ► The probe depletion method abrogating the need for fluorophore labeled in capillary. ► The probe depletion approach is applicable to both RNA and DNA viruses. ► The working principle may be applied in miniaturized portable devices in the future.