| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 4360817 | Cell Host & Microbe | 2016 | 8 Pages |
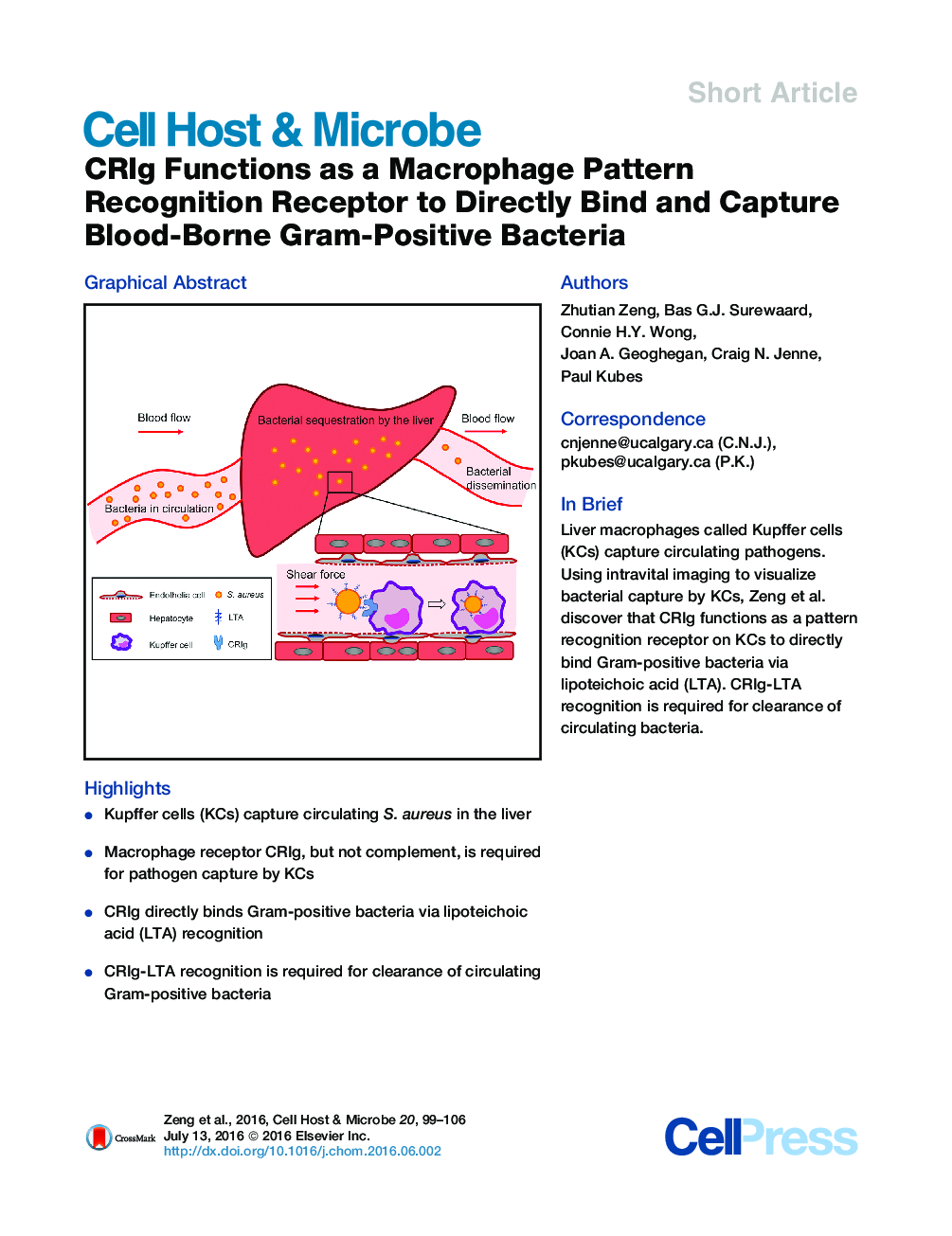
•Kupffer cells (KCs) capture circulating S. aureus in the liver•Macrophage receptor CRIg, but not complement, is required for pathogen capture by KCs•CRIg directly binds Gram-positive bacteria via lipoteichoic acid (LTA) recognition•CRIg-LTA recognition is required for clearance of circulating Gram-positive bacteria
SummaryKupffer cells (KCs), the vast pool of intravascular macrophages in the liver, help to clear blood-borne pathogens. The mechanisms by which KCs capture circulating pathogens remain unknown. Here we use intra-vital imaging of mice infected with Staphylococcus aureus to directly visualize the dynamic process of bacterial capture in the liver. Circulating S. aureus were captured by KCs in a manner dependent on the macrophage complement receptor CRIg, but the process was independent of complement. CRIg bound Staphylococcus aureus specifically through recognition of lipoteichoic acid (LTA), but not cell-wall-anchored surface proteins or peptidoglycan. Blocking the recognition between CRIg and LTA in vivo diminished the bacterial capture in liver and led to systemic bacterial dissemination. All tested Gram-positive, but not Gram-negative, bacteria bound CRIg in a complement-independent manner. These findings reveal a pattern recognition role for CRIg in the direct capture of circulating Gram-positive bacteria from the bloodstream.
Graphical AbstractFigure optionsDownload full-size imageDownload high-quality image (187 K)Download as PowerPoint slide