| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 606442 | Journal of Colloid and Interface Science | 2016 | 10 Pages |
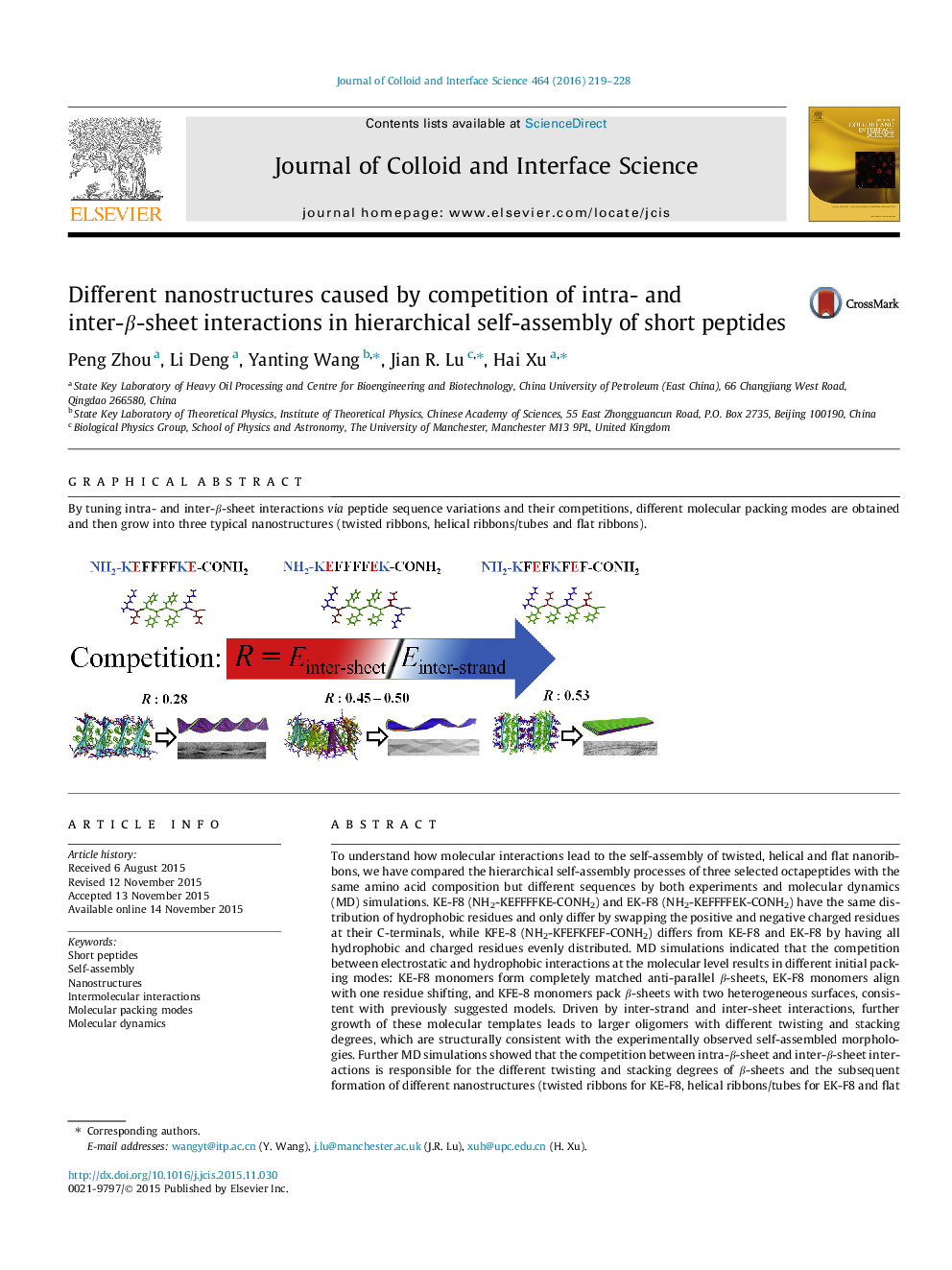
To understand how molecular interactions lead to the self-assembly of twisted, helical and flat nanoribbons, we have compared the hierarchical self-assembly processes of three selected octapeptides with the same amino acid composition but different sequences by both experiments and molecular dynamics (MD) simulations. KE-F8 (NH2-KEFFFFKE-CONH2) and EK-F8 (NH2-KEFFFFEK-CONH2) have the same distribution of hydrophobic residues and only differ by swapping the positive and negative charged residues at their C-terminals, while KFE-8 (NH2-KFEFKFEF-CONH2) differs from KE-F8 and EK-F8 by having all hydrophobic and charged residues evenly distributed. MD simulations indicated that the competition between electrostatic and hydrophobic interactions at the molecular level results in different initial packing modes: KE-F8 monomers form completely matched anti-parallel β-sheets, EK-F8 monomers align with one residue shifting, and KFE-8 monomers pack β-sheets with two heterogeneous surfaces, consistent with previously suggested models. Driven by inter-strand and inter-sheet interactions, further growth of these molecular templates leads to larger oligomers with different twisting and stacking degrees, which are structurally consistent with the experimentally observed self-assembled morphologies. Further MD simulations showed that the competition between intra-β-sheet and inter-β-sheet interactions is responsible for the different twisting and stacking degrees of β-sheets and the subsequent formation of different nanostructures (twisted ribbons for KE-F8, helical ribbons/tubes for EK-F8 and flat ribbons for KFE-8). This study thus provided an important mechanistic insight into the fine tuning of molecular packing and interactions via peptide sequence variation leading to controllable self-assembly of twisted, helical and flat nanostructures.
Graphical abstractBy tuning intra- and inter-β-sheet interactions via peptide sequence variations and their competitions, different molecular packing modes are obtained and then grow into three typical nanostructures (twisted ribbons, helical ribbons/tubes and flat ribbons).Figure optionsDownload full-size imageDownload high-quality image (238 K)Download as PowerPoint slide