| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 6369525 | Journal of Theoretical Biology | 2015 | 10 Pages |
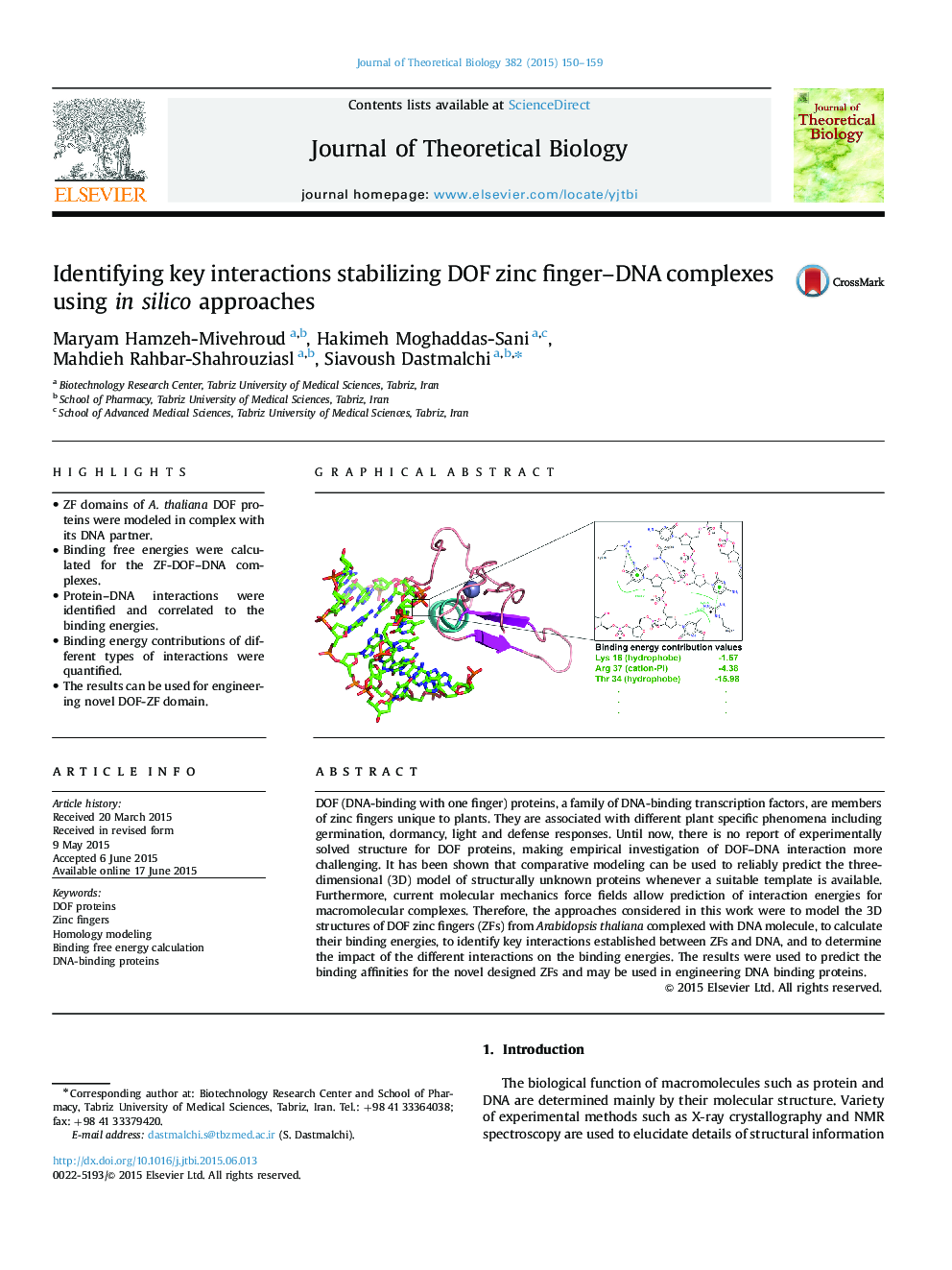
â¢ZF domains of A. thaliana DOF proteins were modeled in complex with its DNA partner.â¢Binding free energies were calculated for the ZF-DOF-DNA complexes.â¢Protein-DNA interactions were identified and correlated to the binding energies.â¢Binding energy contributions of different types of interactions were quantified.â¢The results can be used for engineering novel DOF-ZF domain.
DOF (DNA-binding with one finger) proteins, a family of DNA-binding transcription factors, are members of zinc fingers unique to plants. They are associated with different plant specific phenomena including germination, dormancy, light and defense responses. Until now, there is no report of experimentally solved structure for DOF proteins, making empirical investigation of DOF-DNA interaction more challenging. It has been shown that comparative modeling can be used to reliably predict the three-dimensional (3D) model of structurally unknown proteins whenever a suitable template is available. Furthermore, current molecular mechanics force fields allow prediction of interaction energies for macromolecular complexes. Therefore, the approaches considered in this work were to model the 3D structures of DOF zinc fingers (ZFs) from Arabidopsis thaliana complexed with DNA molecule, to calculate their binding energies, to identify key interactions established between ZFs and DNA, and to determine the impact of the different interactions on the binding energies. The results were used to predict the binding affinities for the novel designed ZFs and may be used in engineering DNA binding proteins.
Graphical abstractDownload full-size image