| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 2039287 | Cell Reports | 2016 | 11 Pages |
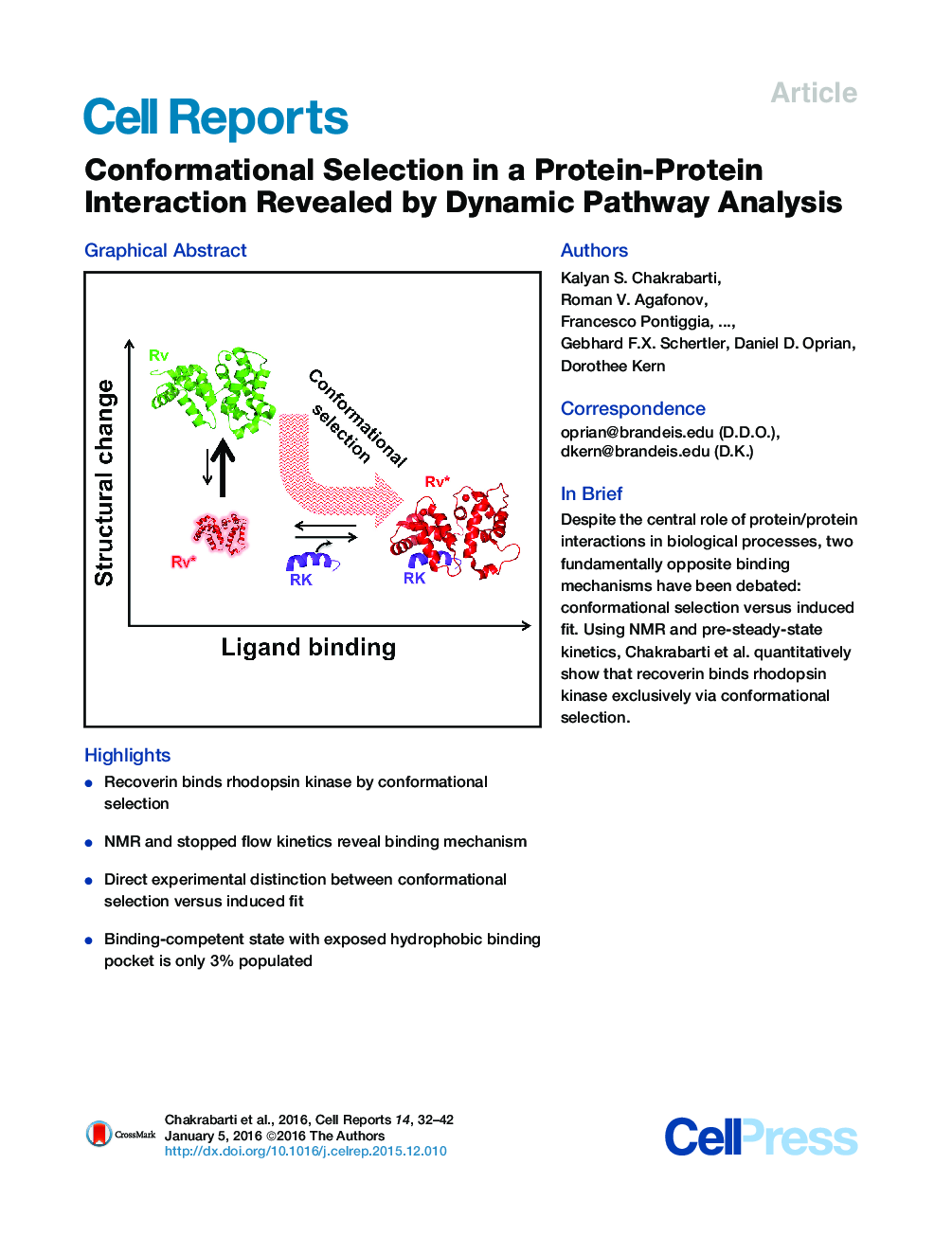
•Recoverin binds rhodopsin kinase by conformational selection•NMR and stopped flow kinetics reveal binding mechanism•Direct experimental distinction between conformational selection versus induced fit•Binding-competent state with exposed hydrophobic binding pocket is only 3% populated
SummaryMolecular recognition plays a central role in biology, and protein dynamics has been acknowledged to be important in this process. However, it is highly debated whether conformational changes happen before ligand binding to produce a binding-competent state (conformational selection) or are caused in response to ligand binding (induced fit). Proposals for both mechanisms in protein/protein recognition have been primarily based on structural arguments. However, the distinction between them is a question of the probabilities of going via these two opposing pathways. Here, we present a direct demonstration of exclusive conformational selection in protein/protein recognition by measuring the flux for rhodopsin kinase binding to its regulator recoverin, an important molecular recognition in the vision system. Using nuclear magnetic resonance (NMR) spectroscopy, stopped-flow kinetics, and isothermal titration calorimetry, we show that recoverin populates a minor conformation in solution that exposes a hydrophobic binding pocket responsible for binding rhodopsin kinase. Protein dynamics in free recoverin limits the overall rate of binding.
Graphical AbstractFigure optionsDownload full-size imageDownload as PowerPoint slide