| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 2039796 | Cell Reports | 2016 | 11 Pages |
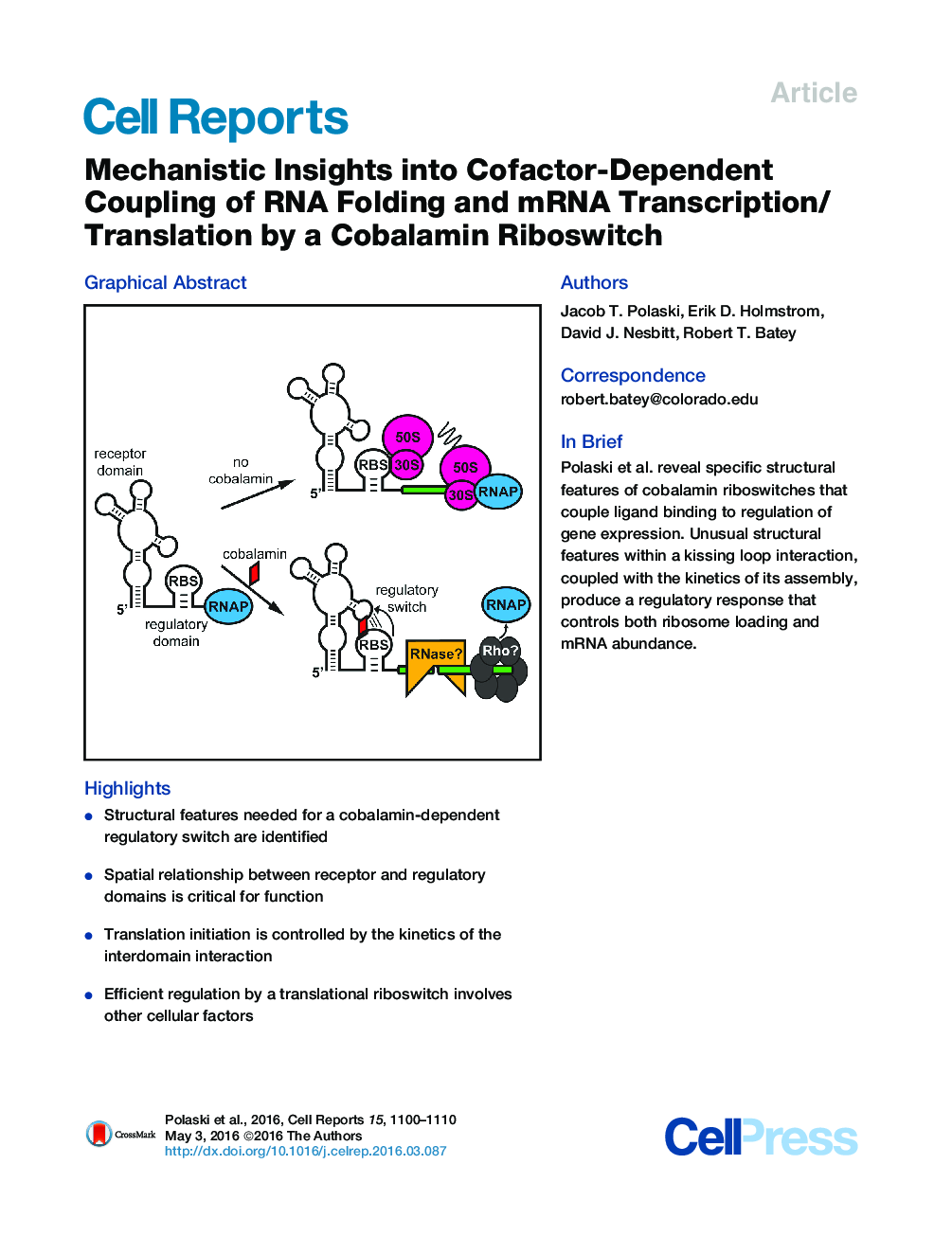
•Structural features needed for a cobalamin-dependent regulatory switch are identified•Spatial relationship between receptor and regulatory domains is critical for function•Translation initiation is controlled by the kinetics of the interdomain interaction•Efficient regulation by a translational riboswitch involves other cellular factors
SummaryRiboswitches are mRNA elements regulating gene expression in response to direct binding of a metabolite. While these RNAs are increasingly well understood with respect to interactions between receptor domains and their cognate effector molecules, little is known about the specific mechanistic relationship between metabolite binding and gene regulation by the downstream regulatory domain. Using a combination of cell-based, biochemical, and biophysical techniques, we reveal the specific RNA architectural features enabling a cobalamin-dependent hairpin loop docking interaction between receptor and regulatory domains. Furthermore, these data demonstrate that docking kinetics dictate a regulatory response involving the coupling of translation initiation to general mechanisms that control mRNA abundance. These results yield a comprehensive picture of how RNA structure in the riboswitch regulatory domain enables kinetically constrained ligand-dependent regulation of gene expression.
Graphical AbstractFigure optionsDownload full-size imageDownload as PowerPoint slide