| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 2039865 | Cell Reports | 2015 | 14 Pages |
•Correlated evolutionary histories predict functions for human genes•Reconstructing shared ancestry extends predictive ability to gene families•Scalable scoring metric leads to unbiased genome-wide functional predictions•Predictions are experimentally tractable, and a subset was validated in cell lines
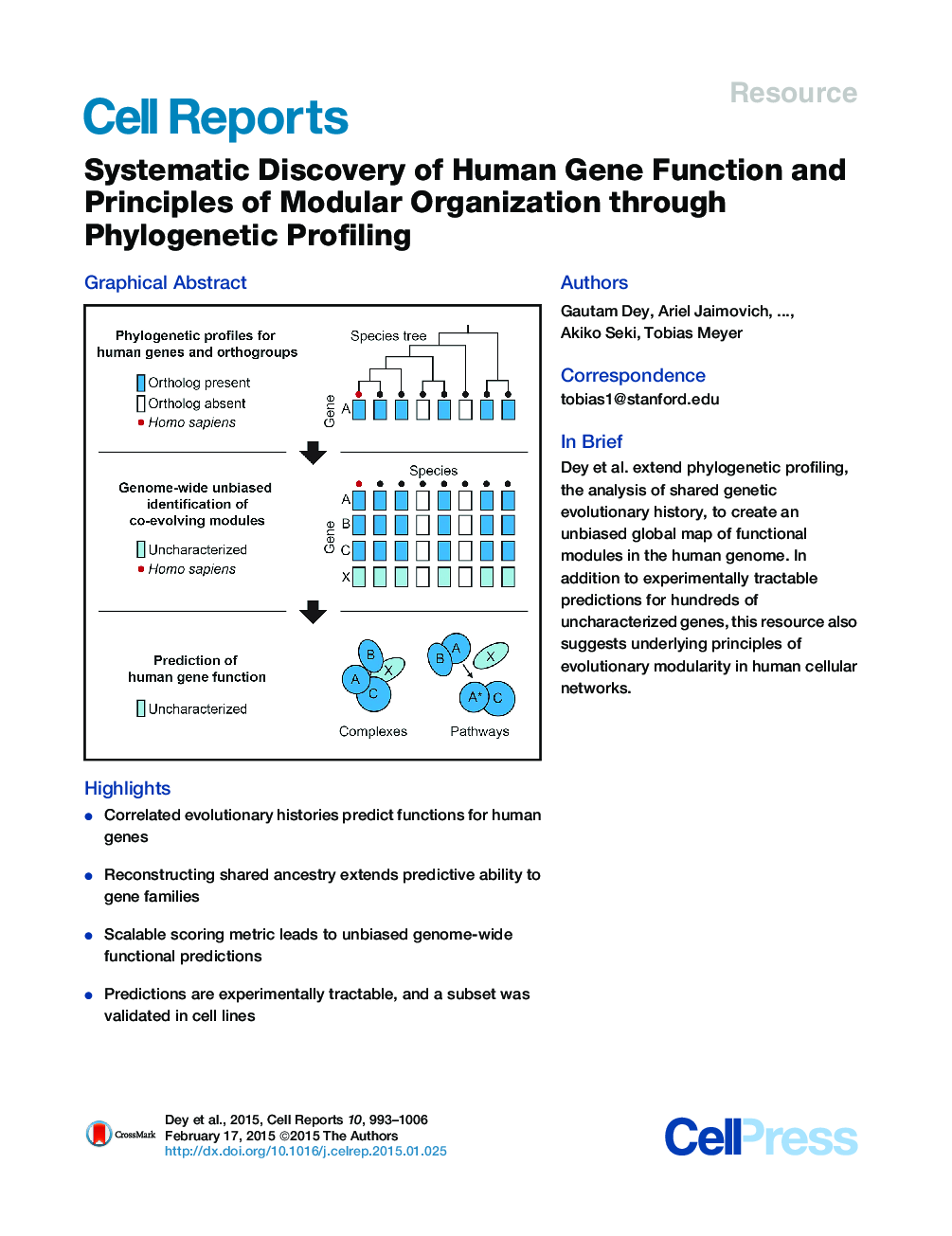
SummaryFunctional links between genes can be predicted using phylogenetic profiling, by correlating the appearance and loss of homologs in subsets of species. However, effective genome-wide phylogenetic profiling has been hindered by the large fraction of human genes related to each other through historical duplication events. Here, we overcame this challenge by automatically profiling over 30,000 groups of homologous human genes (orthogroups) representing the entire protein-coding genome across 177 eukaryotic species (hOP profiles). By generating a full pairwise orthogroup phylogenetic co-occurrence matrix, we derive unbiased genome-wide predictions of functional modules (hOP modules). Our approach predicts functions for hundreds of poorly characterized genes. The results suggest evolutionary constraints that lead components of protein complexes and metabolic pathways to co-evolve while genes in signaling and transcriptional networks do not. As a proof of principle, we validated two subsets of candidates experimentally for their predicted link to the actin-nucleating WASH complex and cilia/basal body function.
Graphical AbstractFigure optionsDownload full-size imageDownload as PowerPoint slide