| Article ID | Journal | Published Year | Pages | File Type |
|---|---|---|---|---|
| 2040418 | Cell Reports | 2016 | 6 Pages |
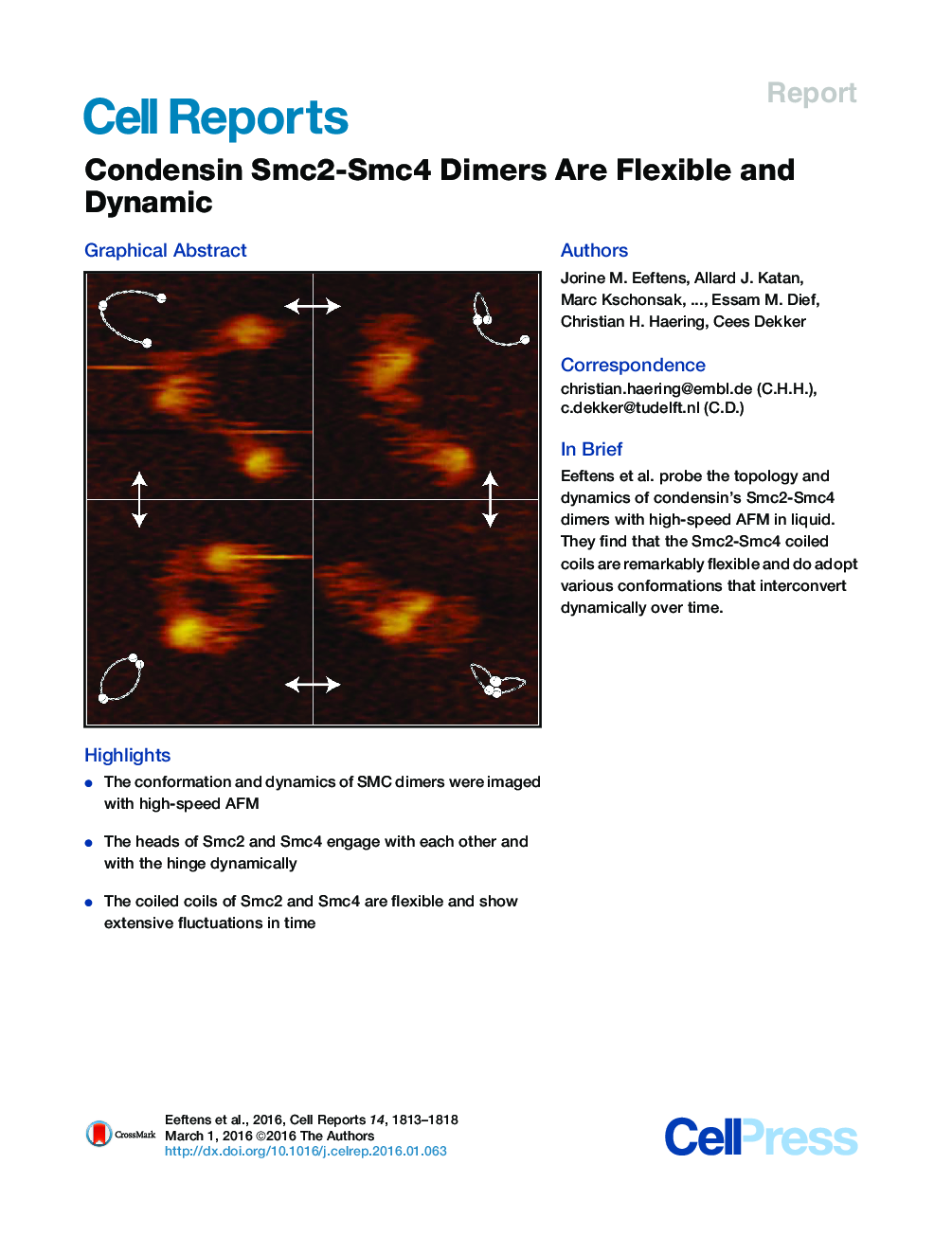
•The conformation and dynamics of SMC dimers were imaged with high-speed AFM•The heads of Smc2 and Smc4 engage with each other and with the hinge dynamically•The coiled coils of Smc2 and Smc4 are flexible and show extensive fluctuations in time
SummaryStructural maintenance of chromosomes (SMC) protein complexes, including cohesin and condensin, play key roles in the regulation of higher-order chromosome organization. Even though SMC proteins are thought to mechanistically determine the function of the complexes, their native conformations and dynamics have remained unclear. Here, we probe the topology of Smc2-Smc4 dimers of the S. cerevisiae condensin complex with high-speed atomic force microscopy (AFM) in liquid. We show that the Smc2-Smc4 coiled coils are highly flexible polymers with a persistence length of only ∼4 nm. Moreover, we demonstrate that the SMC dimers can adopt various architectures that interconvert dynamically over time, and we find that the SMC head domains engage not only with each other, but also with the hinge domain situated at the other end of the ∼45-nm-long coiled coil. Our findings reveal structural properties that provide insights into the molecular mechanics of condensin complexes.
Graphical AbstractFigure optionsDownload full-size imageDownload as PowerPoint slide