| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 1229606 | 1495236 | 2014 | 13 صفحه PDF | دانلود رایگان |
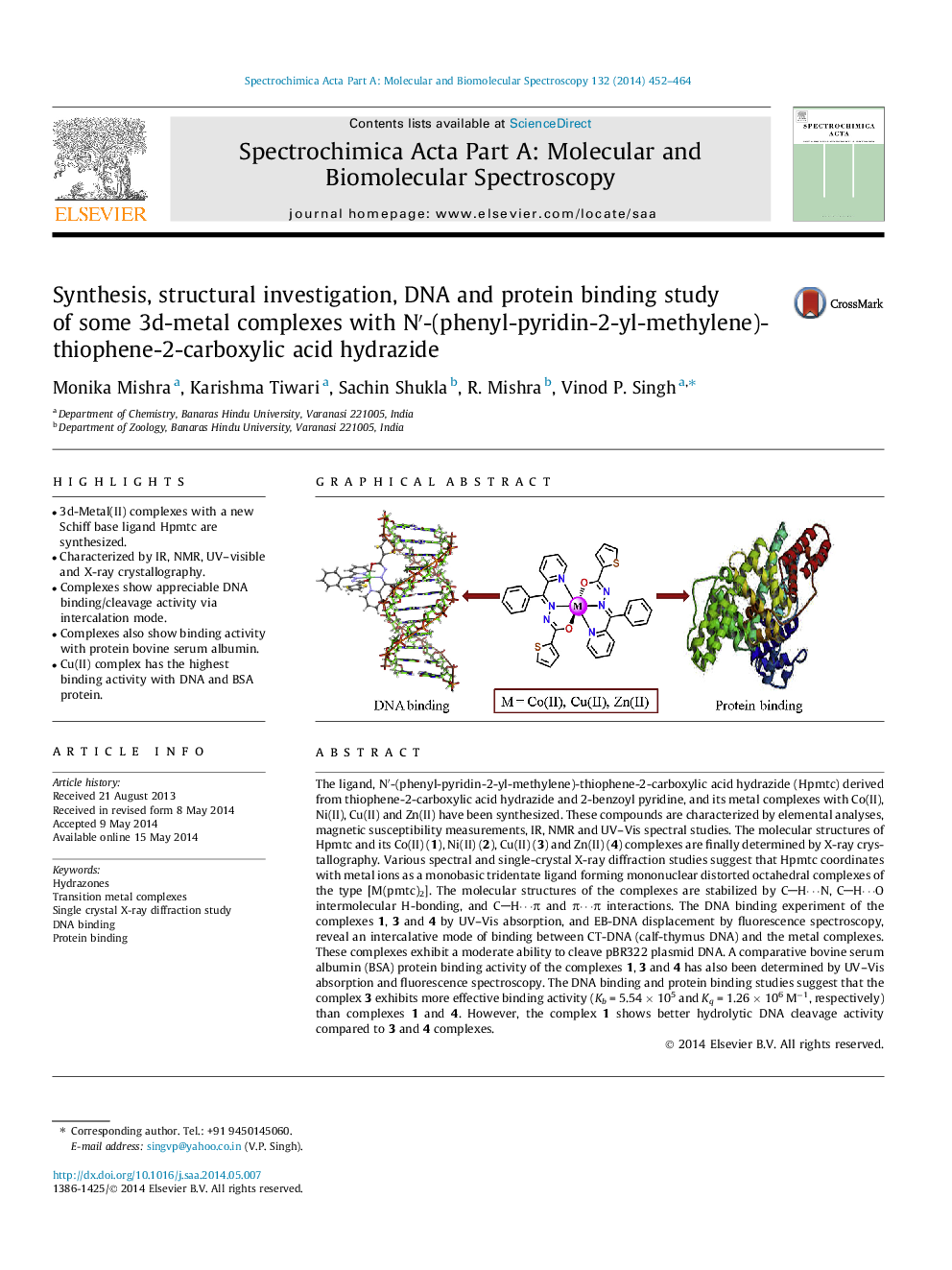
• 3d-Metal(II) complexes with a new Schiff base ligand Hpmtc are synthesized.
• Characterized by IR, NMR, UV–visible and X-ray crystallography.
• Complexes show appreciable DNA binding/cleavage activity via intercalation mode.
• Complexes also show binding activity with protein bovine serum albumin.
• Cu(II) complex has the highest binding activity with DNA and BSA protein.
The ligand, N′-(phenyl-pyridin-2-yl-methylene)-thiophene-2-carboxylic acid hydrazide (Hpmtc) derived from thiophene-2-carboxylic acid hydrazide and 2-benzoyl pyridine, and its metal complexes with Co(II), Ni(II), Cu(II) and Zn(II) have been synthesized. These compounds are characterized by elemental analyses, magnetic susceptibility measurements, IR, NMR and UV–Vis spectral studies. The molecular structures of Hpmtc and its Co(II) (1), Ni(II) (2), Cu(II) (3) and Zn(II) (4) complexes are finally determined by X-ray crystallography. Various spectral and single-crystal X-ray diffraction studies suggest that Hpmtc coordinates with metal ions as a monobasic tridentate ligand forming mononuclear distorted octahedral complexes of the type [M(pmtc)2]. The molecular structures of the complexes are stabilized by CH⋯N, CH⋯O intermolecular H-bonding, and CH⋯π and π⋯π interactions. The DNA binding experiment of the complexes 1, 3 and 4 by UV–Vis absorption, and EB-DNA displacement by fluorescence spectroscopy, reveal an intercalative mode of binding between CT-DNA (calf-thymus DNA) and the metal complexes. These complexes exhibit a moderate ability to cleave pBR322 plasmid DNA. A comparative bovine serum albumin (BSA) protein binding activity of the complexes 1, 3 and 4 has also been determined by UV–Vis absorption and fluorescence spectroscopy. The DNA binding and protein binding studies suggest that the complex 3 exhibits more effective binding activity (Kb = 5.54 × 105 and Kq = 1.26 × 106 M−1, respectively) than complexes 1 and 4. However, the complex 1 shows better hydrolytic DNA cleavage activity compared to 3 and 4 complexes.
Figure optionsDownload as PowerPoint slide
Journal: Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy - Volume 132, 11 November 2014, Pages 452–464