| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 2039116 | 1073027 | 2015 | 13 صفحه PDF | دانلود رایگان |
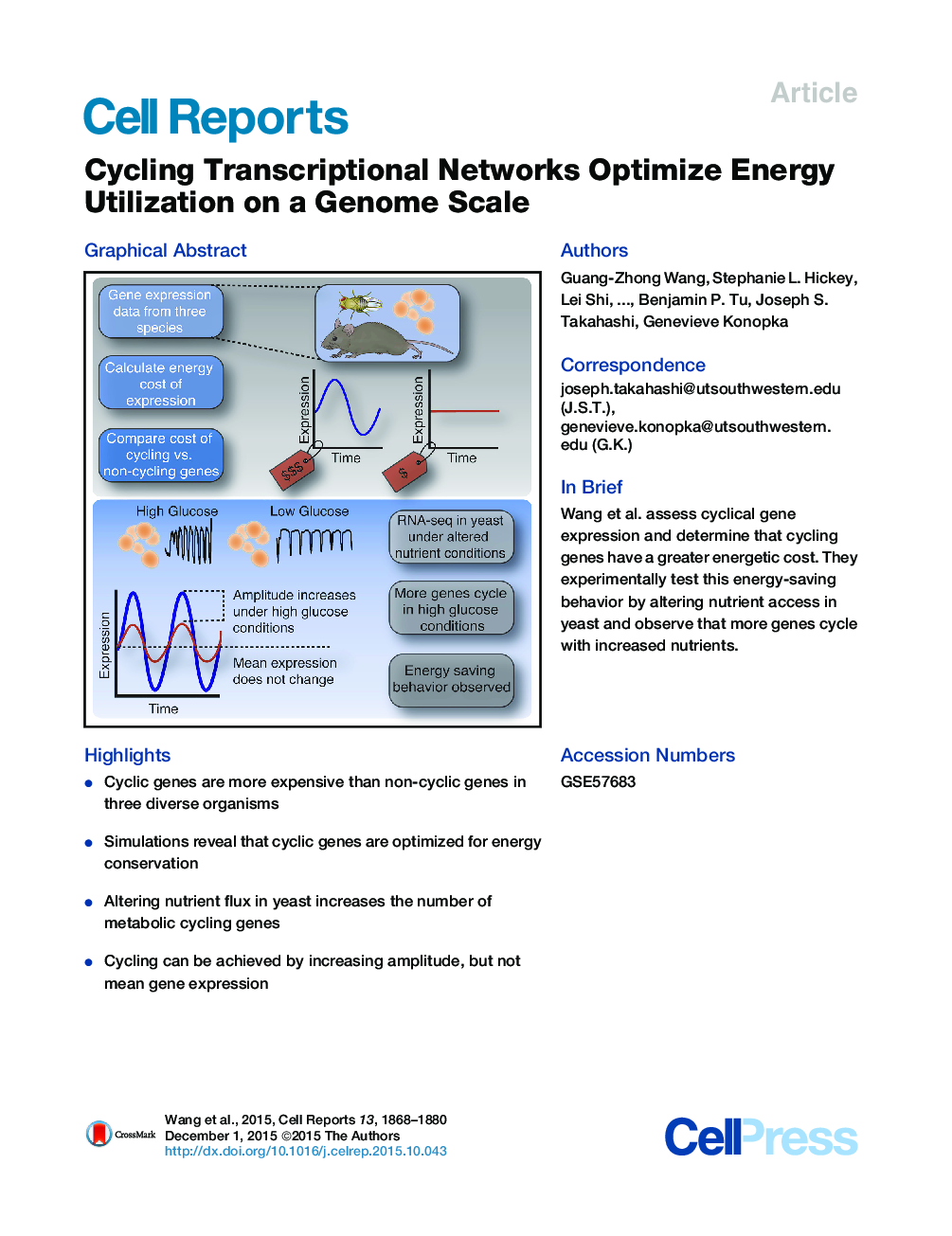
• Cyclic genes are more expensive than non-cyclic genes in three diverse organisms
• Simulations reveal that cyclic genes are optimized for energy conservation
• Altering nutrient flux in yeast increases the number of metabolic cycling genes
• Cycling can be achieved by increasing amplitude, but not mean gene expression
SummaryGenes expressing circadian RNA rhythms are enriched for metabolic pathways, but the adaptive significance of cyclic gene expression remains unclear. We estimated the genome-wide synthetic and degradative cost of transcription and translation in three organisms and found that the cost of cycling genes is strikingly higher compared to non-cycling genes. Cycling genes are expressed at high levels and constitute the most costly proteins to synthesize in the genome. We demonstrate that metabolic cycling is accelerated in yeast grown under higher nutrient flux and the number of cycling genes increases ∼40%, which are achieved by increasing the amplitude and not the mean level of gene expression. These results suggest that rhythmic gene expression optimizes the metabolic cost of global gene expression and that highly expressed genes have been selected to be downregulated in a cyclic manner for energy conservation.
Graphical AbstractFigure optionsDownload as PowerPoint slide
Journal: - Volume 13, Issue 9, 1 December 2015, Pages 1868–1880