| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 2040067 | 1073095 | 2014 | 12 صفحه PDF | دانلود رایگان |
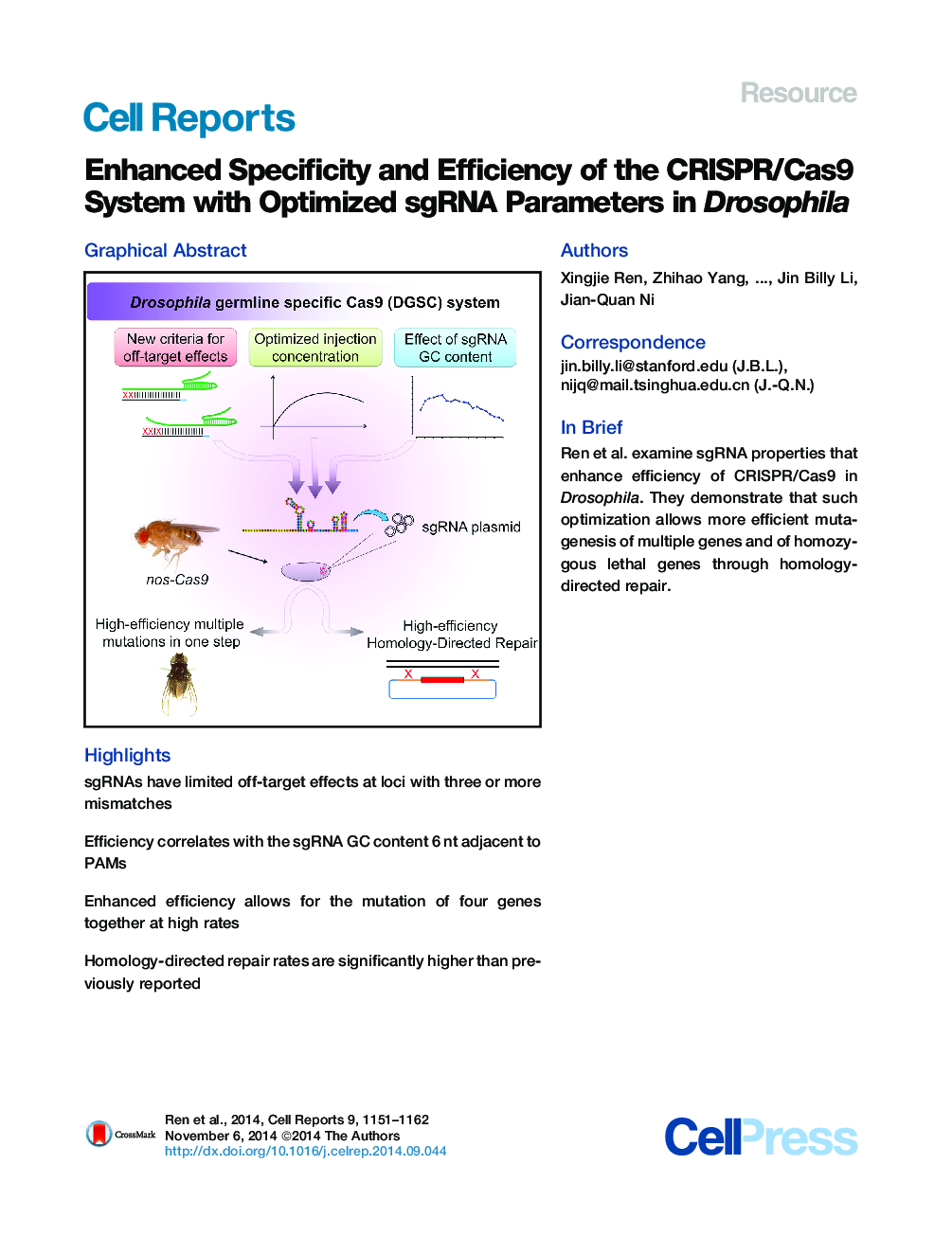
• sgRNAs have limited off-target effects at loci with three or more mismatches
• Efficiency correlates with the sgRNA GC content 6 nt adjacent to PAMs
• Enhanced efficiency allows for the mutation of four genes together at high rates
• Homology-directed repair rates are significantly higher than previously reported
SummaryThe CRISPR/Cas9 system has recently emerged as a powerful tool for functional genomic studies in Drosophila melanogaster. However, single-guide RNA (sgRNA) parameters affecting the specificity and efficiency of the system in flies are still not clear. Here, we found that off-target effects did not occur in regions of genomic DNA with three or more nucleotide mismatches to sgRNAs. Importantly, we document for a strong positive correlation between mutagenesis efficiency and sgRNA GC content of the six protospacer-adjacent motif-proximal nucleotides (PAMPNs). Furthermore, by injecting well-designed sgRNA plasmids at the optimal concentration we determined, we could efficiently generate mutations in four genes in one step. Finally, we generated null alleles of HP1a using optimized parameters through homology-directed repair and achieved an overall mutagenesis rate significantly higher than previously reported. Our work demonstrates a comprehensive optimization of sgRNA and promises to vastly simplify CRISPR/Cas9 experiments in Drosophila.
Graphical AbstractFigure optionsDownload as PowerPoint slide
Journal: - Volume 9, Issue 3, 6 November 2014, Pages 1151–1162