| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 2035351 | 1543093 | 2015 | 13 صفحه PDF | دانلود رایگان |
• Ligand-driven re-orientation of receptor dimer topology tunes signaling output
• Diabodies elicit differential signal activation
• Non-agonistic diabodies counteract intracellular oncogenic signaling
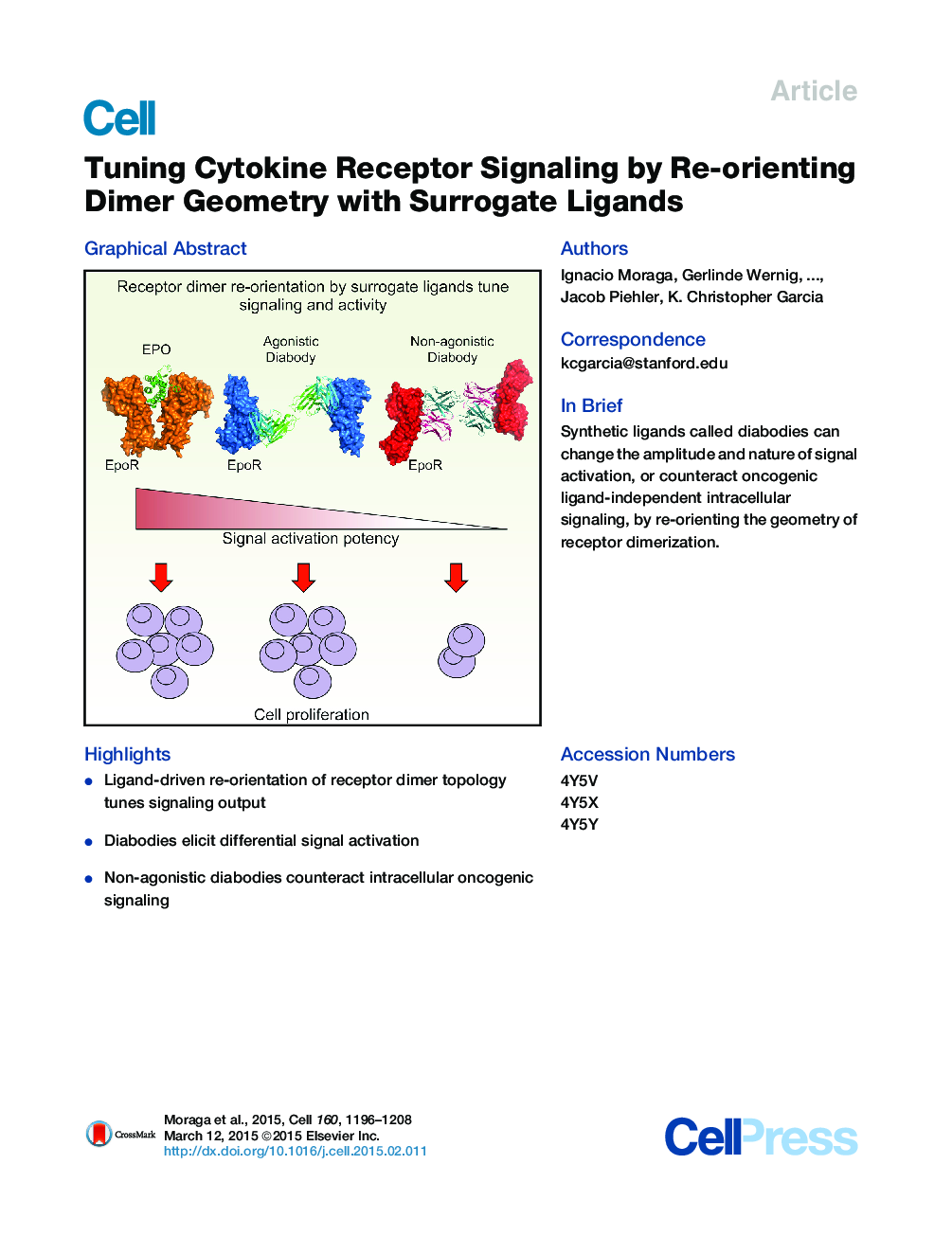
SummaryMost cell-surface receptors for cytokines and growth factors signal as dimers, but it is unclear whether remodeling receptor dimer topology is a viable strategy to “tune” signaling output. We utilized diabodies (DA) as surrogate ligands in a prototypical dimeric receptor-ligand system, the cytokine Erythropoietin (EPO) and its receptor (EpoR), to dimerize EpoR ectodomains in non-native architectures. Diabody-induced signaling amplitudes varied from full to minimal agonism, and structures of these DA/EpoR complexes differed in EpoR dimer orientation and proximity. Diabodies also elicited biased or differential activation of signaling pathways and gene expression profiles compared to EPO. Non-signaling diabodies inhibited proliferation of erythroid precursors from patients with a myeloproliferative neoplasm due to a constitutively active JAK2V617F mutation. Thus, intracellular oncogenic mutations causing ligand-independent receptor activation can be counteracted by extracellular ligands that re-orient receptors into inactive dimer topologies. This approach has broad applications for tuning signaling output for many dimeric receptor systems.
Graphical AbstractFigure optionsDownload high-quality image (236 K)Download as PowerPoint slide
Journal: - Volume 160, Issue 6, 12 March 2015, Pages 1196–1208