| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 4496431 | 1623884 | 2013 | 7 صفحه PDF | دانلود رایگان |
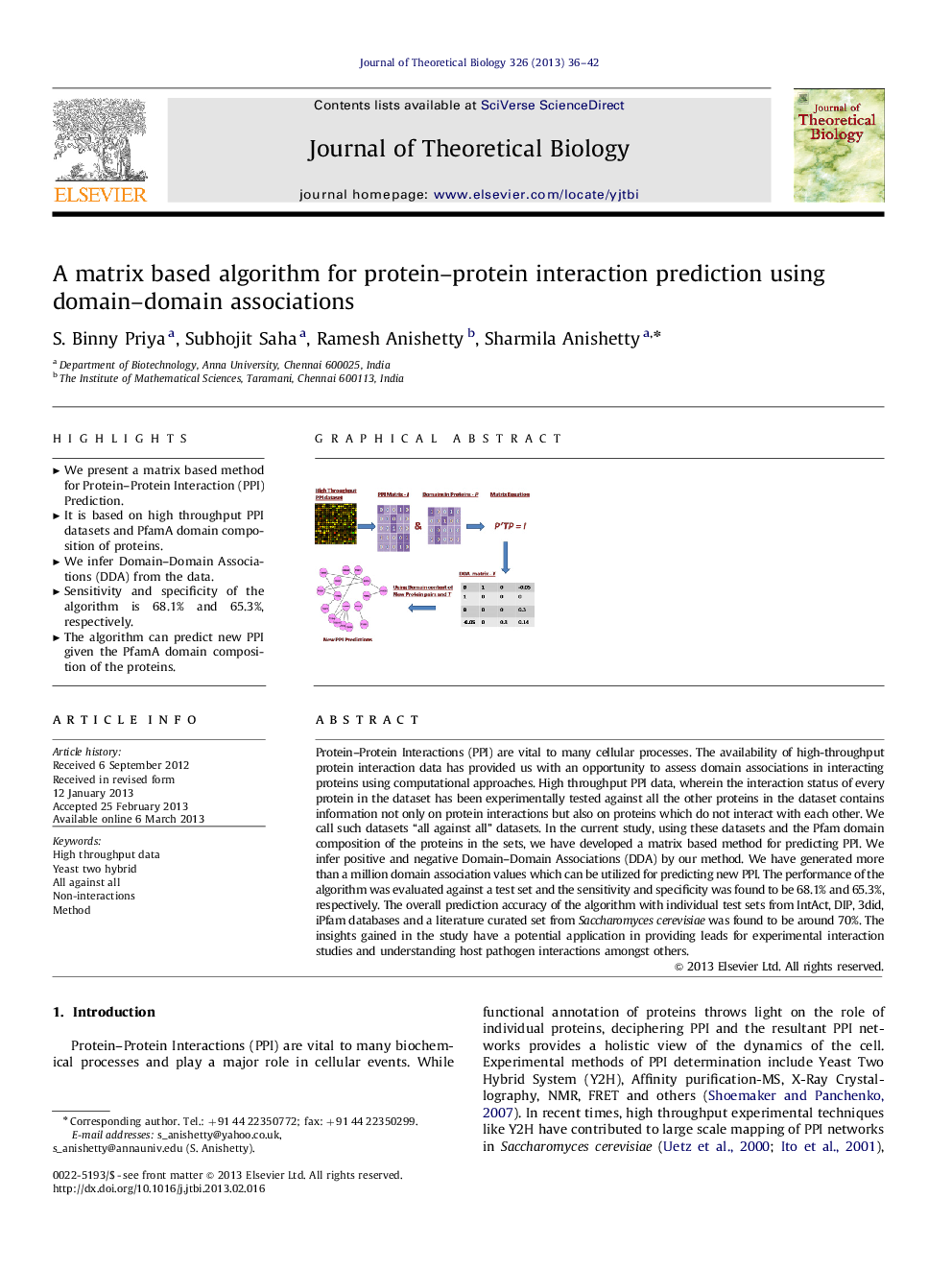
Protein–Protein Interactions (PPI) are vital to many cellular processes. The availability of high-throughput protein interaction data has provided us with an opportunity to assess domain associations in interacting proteins using computational approaches. High throughput PPI data, wherein the interaction status of every protein in the dataset has been experimentally tested against all the other proteins in the dataset contains information not only on protein interactions but also on proteins which do not interact with each other. We call such datasets “all against all” datasets. In the current study, using these datasets and the Pfam domain composition of the proteins in the sets, we have developed a matrix based method for predicting PPI. We infer positive and negative Domain–Domain Associations (DDA) by our method. We have generated more than a million domain association values which can be utilized for predicting new PPI. The performance of the algorithm was evaluated against a test set and the sensitivity and specificity was found to be 68.1% and 65.3%, respectively. The overall prediction accuracy of the algorithm with individual test sets from IntAct, DIP, 3did, iPfam databases and a literature curated set from Saccharomyces cerevisiae was found to be around 70%. The insights gained in the study have a potential application in providing leads for experimental interaction studies and understanding host pathogen interactions amongst others.
Figure optionsDownload as PowerPoint slideHighlights
► We present a matrix based method for Protein–Protein Interaction (PPI) Prediction.
► It is based on high throughput PPI datasets and PfamA domain composition of proteins.
► We infer Domain–Domain Associations (DDA) from the data.
► Sensitivity and specificity of the algorithm is 68.1% and 65.3%, respectively.
► The algorithm can predict new PPI given the PfamA domain composition of the proteins.
Journal: Journal of Theoretical Biology - Volume 326, 7 June 2013, Pages 36–42