| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 1391979 | 983689 | 2015 | 9 صفحه PDF | دانلود رایگان |
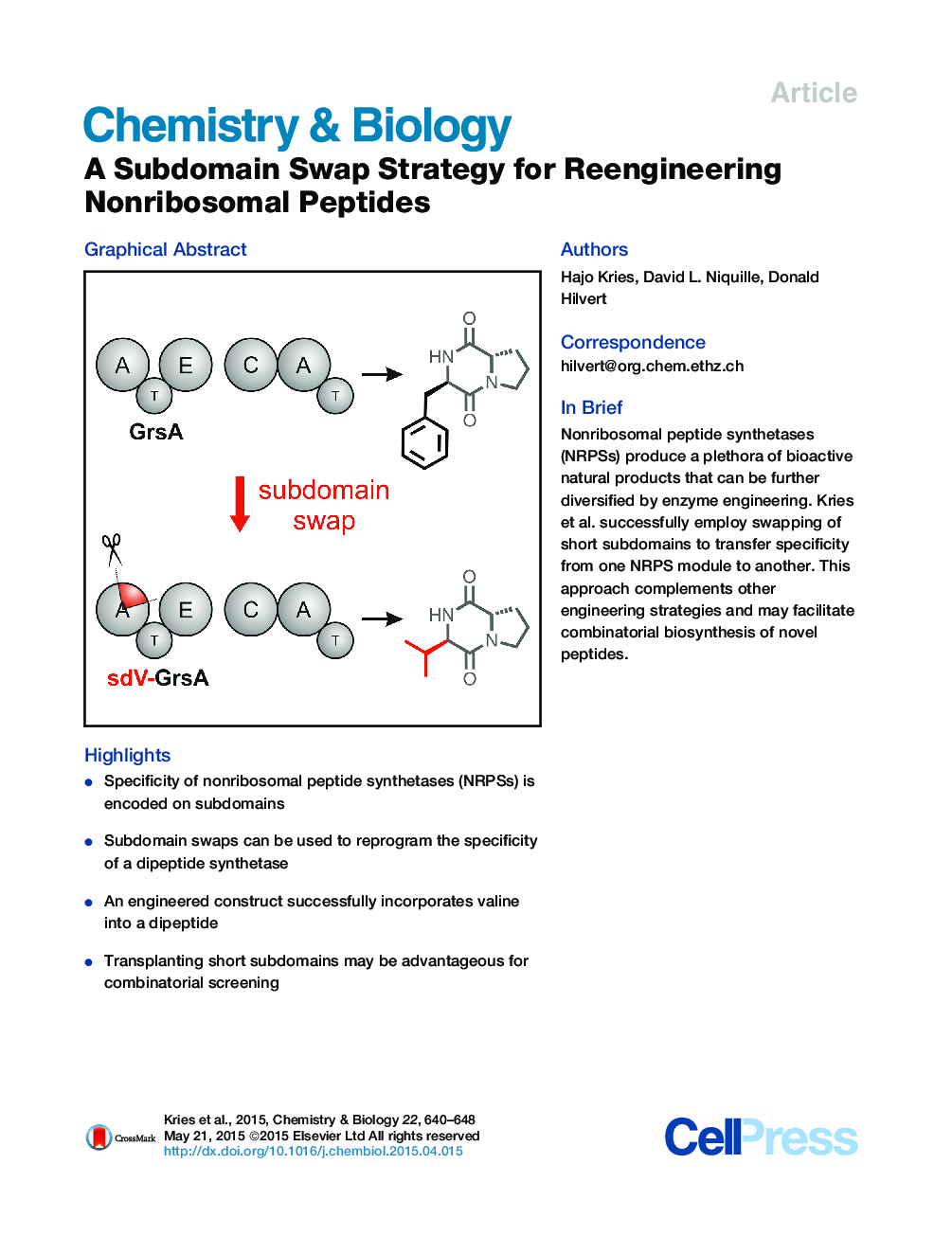
• Specificity of nonribosomal peptide synthetases (NRPSs) is encoded on subdomains
• Subdomain swaps can be used to reprogram the specificity of a dipeptide synthetase
• An engineered construct successfully incorporates valine into a dipeptide
• Transplanting short subdomains may be advantageous for combinatorial screening
SummaryNonribosomal peptide synthetases (NRPSs) protect microorganisms from environmental threats by producing diverse siderophores, antibiotics, and other peptide natural products. Their modular molecular structure is also attractive from the standpoint of biosynthetic engineering. Here we evaluate a methodology for swapping module specificities of these mega-enzymes that takes advantage of flavodoxin-like subdomains involved in substrate recognition. Nine subdomains encoding diverse specificities were transplanted into the Phe-specific GrsA initiation module of gramicidin S synthetase. All chimeras could be purified as soluble protein. One construct based on a Val-specific subdomain showed sizable adenylation activity and functioned as a Val-Pro diketopiperazine synthetase upon addition of the proline-specific GrsB1 module. These results suggest that subdomain swapping could be a viable alternative to previous NRPS design approaches targeting binding pockets, domains, or entire modules. The short length of the swapped sequence stretch may facilitate straightforward exploitation of the wealth of existing NRPS modules for combinatorial biosynthesis.
Graphical AbstractFigure optionsDownload high-quality image (108 K)Download as PowerPoint slide
Journal: - Volume 22, Issue 5, 21 May 2015, Pages 640–648