| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 2029618 | 1070930 | 2016 | 13 صفحه PDF | دانلود رایگان |
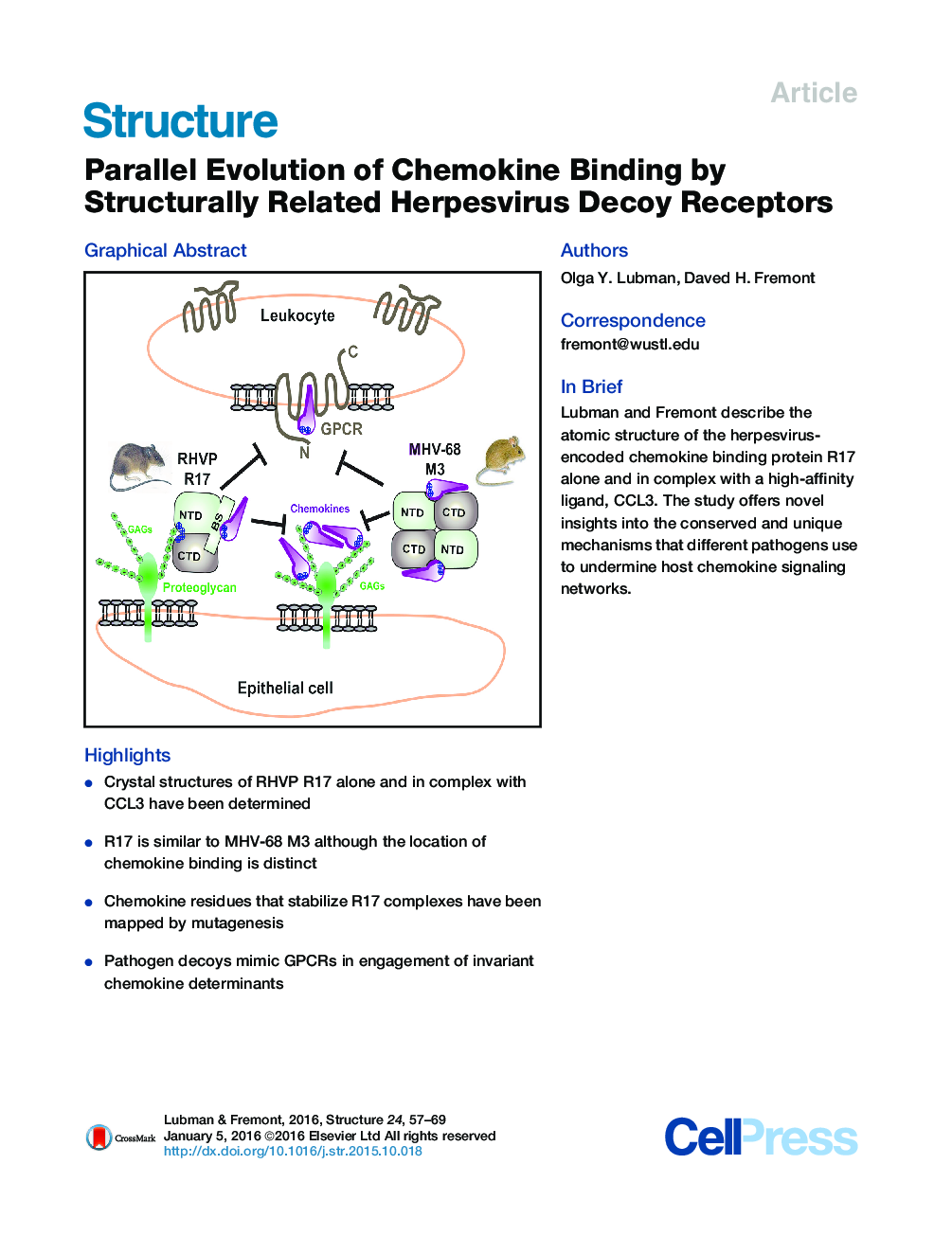
• Crystal structures of RHVP R17 alone and in complex with CCL3 have been determined
• R17 is similar to MHV-68 M3 although the location of chemokine binding is distinct
• Chemokine residues that stabilize R17 complexes have been mapped by mutagenesis
• Pathogen decoys mimic GPCRs in engagement of invariant chemokine determinants
SummaryA wide variety of pathogens targets chemokine signaling networks in order to disrupt host immune surveillance and defense. Here, we report a structural and mutational analysis of rodent herpesvirus Peru encoded R17, a potent chemokine inhibitor that sequesters CC and C chemokines with high affinity. R17 consists of a pair of β-sandwich domains linked together by a bridging sheet, which form an acidic binding cleft for the chemokine CCL3 on the opposite face of a basic surface cluster that binds glycosaminoglycans. R17 promiscuously engages chemokines primarily through the same N-loop determinants used for host receptor recognition while residues located in the chemokine 40s loop drive kinetically stable complex formation. The core fold adopted by R17 is unexpectedly similar to that of the M3 chemokine decoy receptor encoded by MHV-68, although, strikingly, neither the location of ligand engagement nor the stoichiometry of binding is conserved, suggesting that their functions evolved independently.
Graphical AbstractFigure optionsDownload high-quality image (208 K)Download as PowerPoint slide
Journal: - Volume 24, Issue 1, 5 January 2016, Pages 57–69