| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 1365281 | 981557 | 2006 | 14 صفحه PDF | دانلود رایگان |
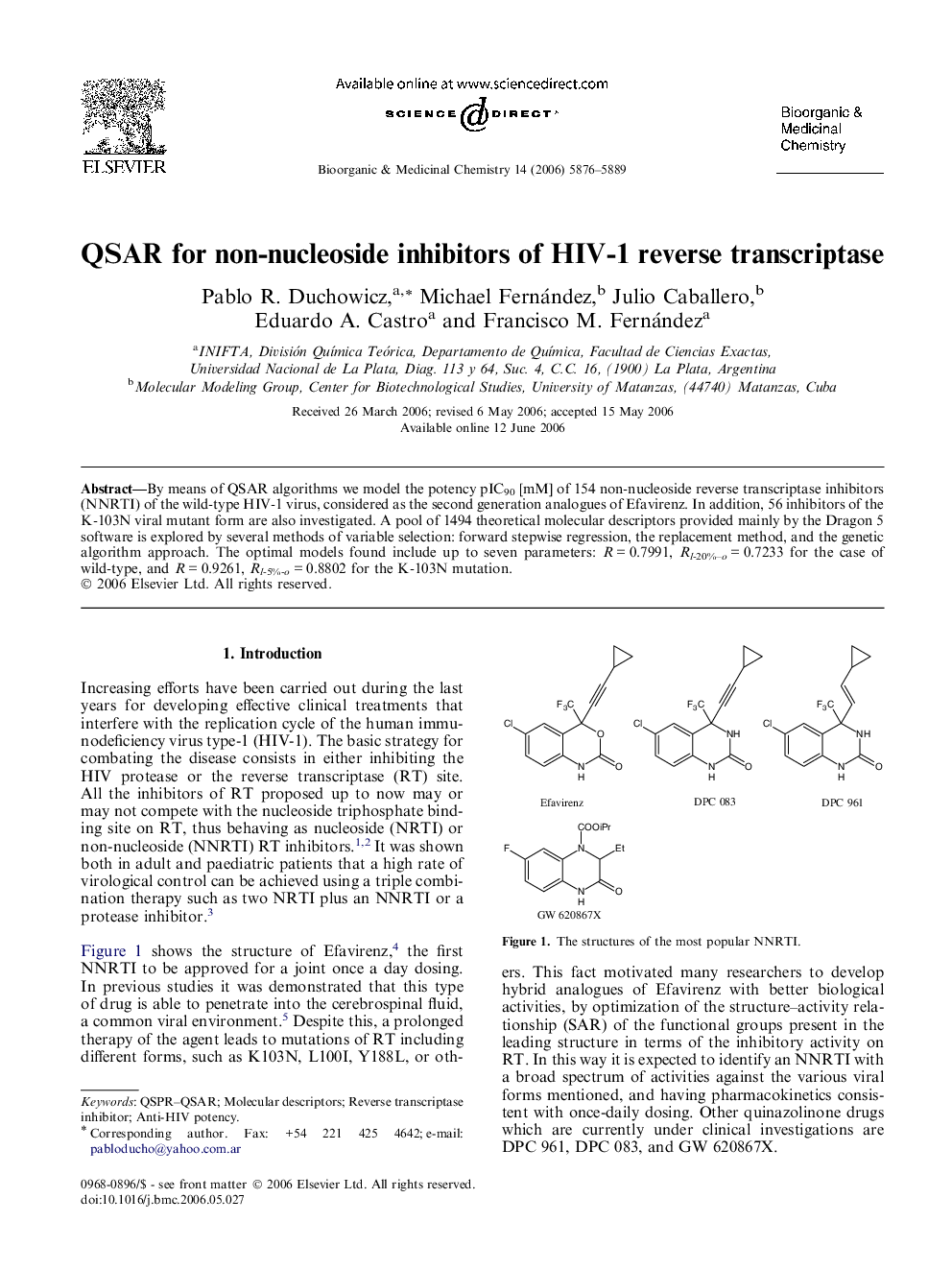
By means of QSAR algorithms we model the potency pIC90 [mM] of 154 non-nucleoside reverse transcriptase inhibitors (NNRTI) of the wild-type HIV-1 virus, considered as the second generation analogues of Efavirenz. In addition, 56 inhibitors of the K-103N viral mutant form are also investigated. A pool of 1494 theoretical molecular descriptors provided mainly by the Dragon 5 software is explored by several methods of variable selection: forward stepwise regression, the replacement method, and the genetic algorithm approach. The optimal models found include up to seven parameters: R = 0.7991, Rl-20%–o = 0.7233 for the case of wild-type, and R = 0.9261, Rl-5%-o = 0.8802 for the K-103N mutation.
QSAR modeling the inhibitory potency pIC90 [mM] of 154 NNRTI of wild-type HIV-1 RT and 56 NNRTI of K-103N mutant form, using forward stepwise regression, the replacement method, and genetic algorithms. Different parallelisms are argued between the activities and the optimal molecular descriptors. Inhibition = function (molecular structural descriptors).Figure optionsDownload as PowerPoint slide
Journal: Bioorganic & Medicinal Chemistry - Volume 14, Issue 17, 1 September 2006, Pages 5876–5889