| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 2937538 | 1576542 | 2016 | 16 صفحه PDF | دانلود رایگان |
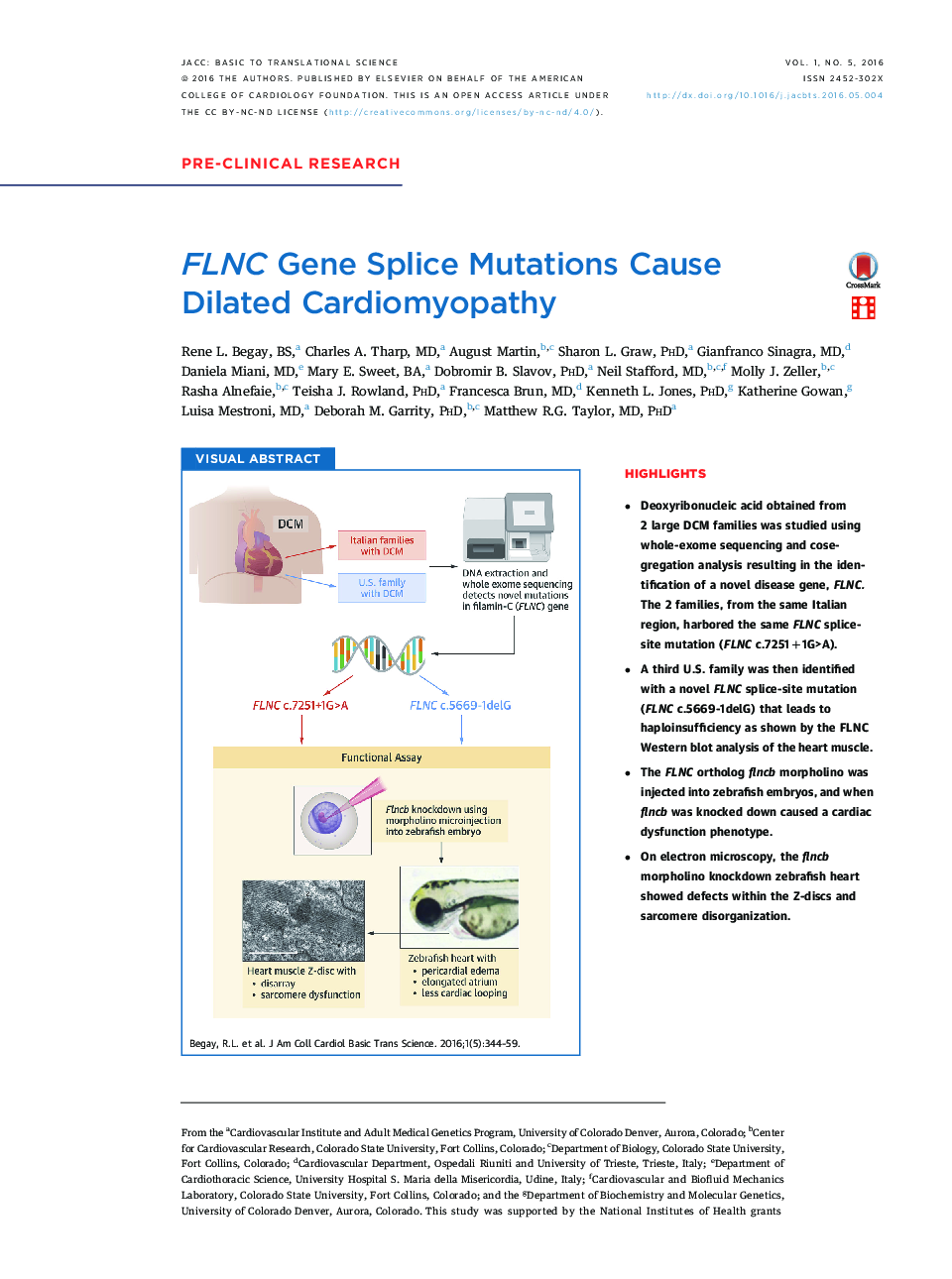
• Deoxyribonucleic acid obtained from 2 large DCM families was studied using whole-exome sequencing and cosegregation analysis resulting in the identification of a novel disease gene, FLNC. The 2 families, from the same Italian region, harbored the same FLNC splice-site mutation (FLNC c.7251+1G>A).
• A third U.S. family was then identified with a novel FLNC splice-site mutation (FLNC c.5669-1delG) that leads to haploinsufficiency as shown by the FLNC Western blot analysis of the heart muscle.
• The FLNC ortholog flncb morpholino was injected into zebrafish embryos, and when flncb was knocked down caused a cardiac dysfunction phenotype.
• On electron microscopy, the flncb morpholino knockdown zebrafish heart showed defects within the Z-discs and sarcomere disorganization.
SummaryA genetic etiology has been identified in 30% to 40% of dilated cardiomyopathy (DCM) patients, yet only 50% of these cases are associated with a known causative gene variant. Thus, in order to understand the pathophysiology of DCM, it is necessary to identify and characterize additional genes. In this study, whole exome sequencing in combination with segregation analysis was used to identify mutations in a novel gene, filamin C (FLNC), resulting in a cardiac-restricted DCM pathology. Here we provide functional data via zebrafish studies and protein analysis to support a model implicating FLNC haploinsufficiency as a mechanism of DCM.
Visual AbstractFigure optionsDownload high-quality image (980 K)Download as PowerPoint slide
Journal: JACC: Basic to Translational Science - Volume 1, Issue 5, August 2016, Pages 344–359