| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 1402375 | 1501743 | 2014 | 8 صفحه PDF | دانلود رایگان |
• 3D-QSAR of CGRP receptor antagonists.
• Interaction comparisons between our most potent compound and olcegepant.
• Revelation of the interaction between our most potent compound and CGRP receptor.
The migraine never fails to afflict individuals in the world that knows no lack of such cases. CGRP (calcitonin gene-related peptide) is found closely related to migraine and olcegepant (BIBN4096) is effective in alleviating the pain. In our work, the combination of ligand- and receptor-based three-dimensional quantitative structure–activity relationship (3D-QSAR) studies along with molecular docking was applied to provide us insights about how urethanamide, pyridine and aspartate and succinate derivatives (novel CGRP receptor antagonists) play a part in inhibiting the activity of CGRP receptor. The optimal CoMSIA model shows the Q2 of 0.505, R2ncv of 0.992 and its accurate predictive ability was confirmed by checking out an independent test set which gave R2pred value of 0.885. Besides, the 3D contour maps help us identify how different groups affect the antagonist activity while connecting to some key positions. In addition, the docking analysis shows the binding site emerging as the distorted “V” shape and including two binding pockets: one of them is hydrophobic, fixing the structural part 3 of compound 80, the other anchors the part 1 of compound 80. The docking analysis also shows the interaction mechanism between compound 80 and CGRP receptor, similar to the interaction between olcegepant and CGRP receptor. The findings derived from this work reveal the mechanism of related antagonists and facilitate the future rational design of novel antagonists with higher potency.
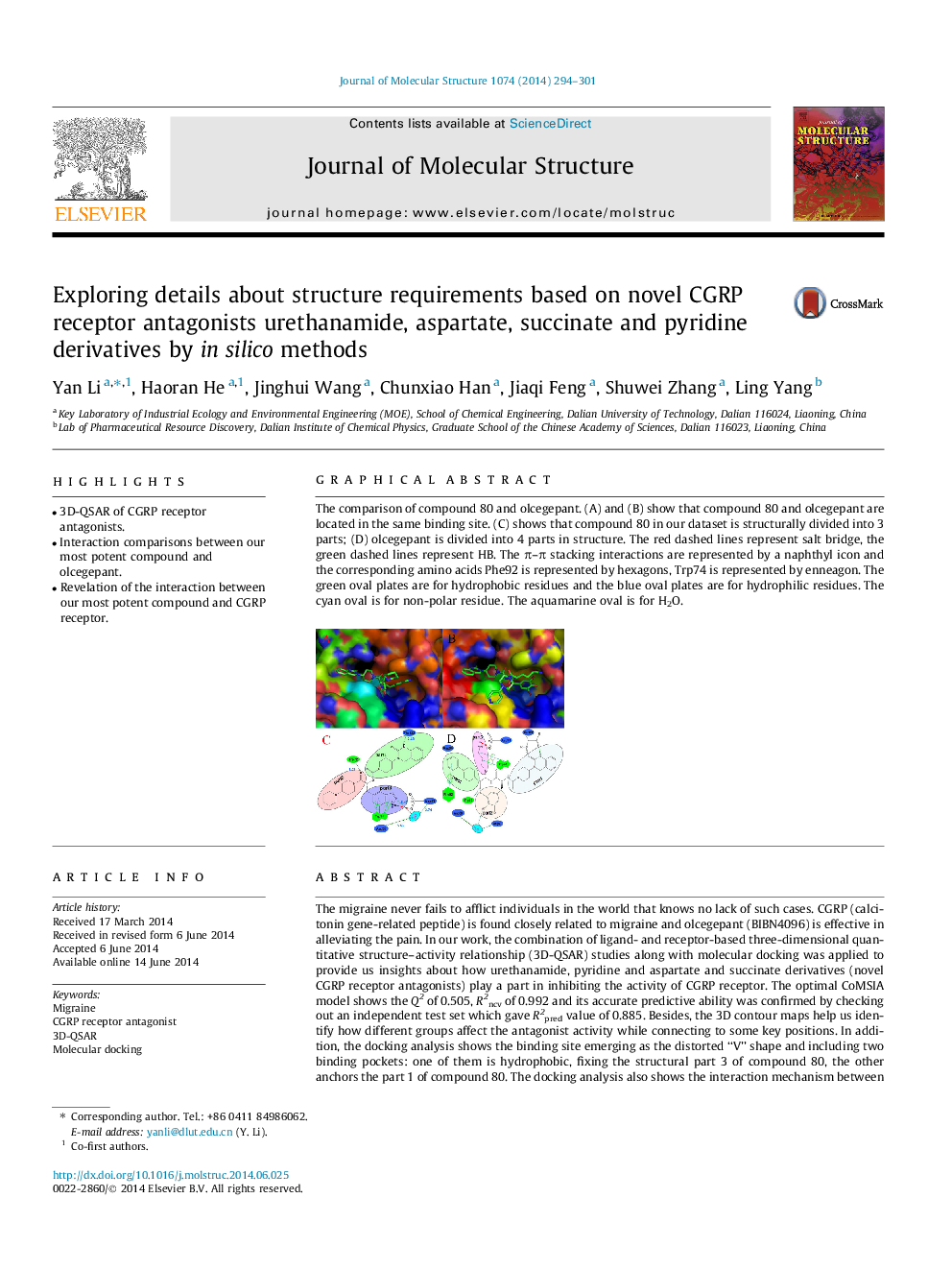
The comparison of compound 80 and olcegepant. (A) and (B) show that compound 80 and olcegepant are located in the same binding site. (C) shows that compound 80 in our dataset is structurally divided into 3 parts; (D) olcegepant is divided into 4 parts in structure. The red dashed lines represent salt bridge, the green dashed lines represent HB. The π–π stacking interactions are represented by a naphthyl icon and the corresponding amino acids Phe92 is represented by hexagons, Trp74 is represented by enneagon. The green oval plates are for hydrophobic residues and the blue oval plates are for hydrophilic residues. The cyan oval is for non-polar residue. The aquamarine oval is for H2O.Figure optionsDownload as PowerPoint slide
Journal: Journal of Molecular Structure - Volume 1074, 25 September 2014, Pages 294–301