| کد مقاله | کد نشریه | سال انتشار | مقاله انگلیسی | نسخه تمام متن |
|---|---|---|---|---|
| 5130787 | 1490856 | 2017 | 10 صفحه PDF | دانلود رایگان |
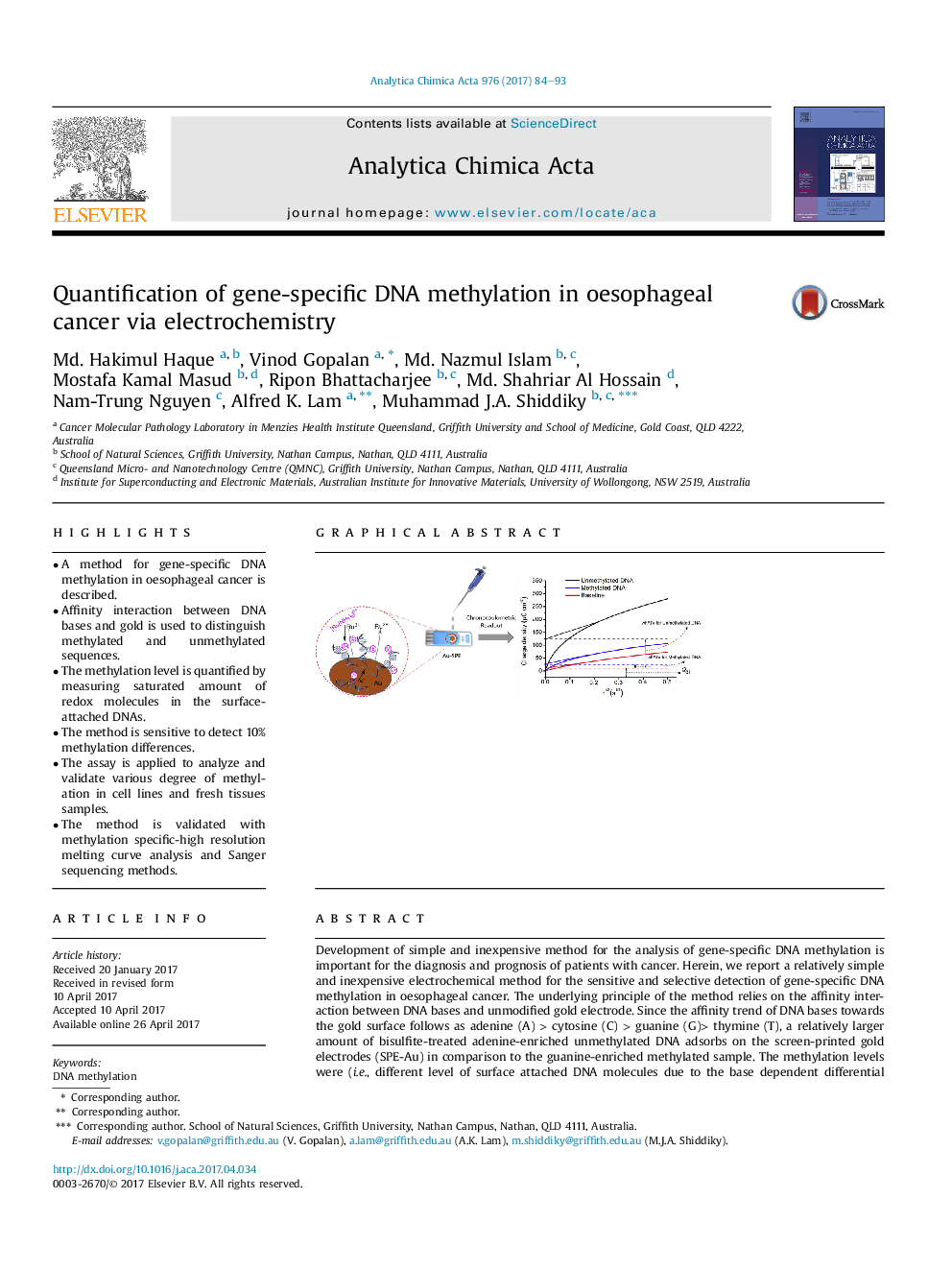
- A method for gene-specific DNA methylation in oesophageal cancer is described.
- Affinity interaction between DNA bases and gold is used to distinguish methylated and unmethylated sequences.
- The methylation level is quantified by measuring saturated amount of redox molecules in the surface-attached DNAs.
- The method is sensitive to detect 10% methylation differences.
- The assay is applied to analyze and validate various degree of methylation in cell lines and fresh tissues samples.
- The method is validated with methylation specific-high resolution melting curve analysis and Sanger sequencing methods.
Development of simple and inexpensive method for the analysis of gene-specific DNA methylation is important for the diagnosis and prognosis of patients with cancer. Herein, we report a relatively simple and inexpensive electrochemical method for the sensitive and selective detection of gene-specific DNA methylation in oesophageal cancer. The underlying principle of the method relies on the affinity interaction between DNA bases and unmodified gold electrode. Since the affinity trend of DNA bases towards the gold surface follows as adenine (A)Â >Â cytosine (C)Â >Â guanine (G)> thymine (T), a relatively larger amount of bisulfite-treated adenine-enriched unmethylated DNA adsorbs on the screen-printed gold electrodes (SPE-Au) in comparison to the guanine-enriched methylated sample. The methylation levels were (i.e., different level of surface attached DNA molecules due to the base dependent differential adsorption pattern) quantified by measuring saturated amount of charge-compensating [Ru(NH3)6]3+ molecules in the surface-attached DNAs by chronocoulometry as redox charge of the [Ru(NH3)6]3+ molecules quantitatively reflects the amount of the adsorbed DNA confined at the electrode surface. The assay could successfully distinguish methylated and unmethylated DNA sequences at single CpG resolution and as low as 10% differences in DNA methylation. In addition, the assay showed fairly good reproducibility (% RSD= <5%) with better sensitivity and specificity by analysing various levels of methylation in two cell lines and eight fresh tissues samples from patients with oesophageal squamous cell carcinoma. Finally, the method was validated with methylation specific-high resolution melting curve analysis and Sanger sequencing methods.
Profiling of gene-specific methylation levels in oesophageal cancer via electrochemistry.227
Journal: Analytica Chimica Acta - Volume 976, 11 July 2017, Pages 84-93